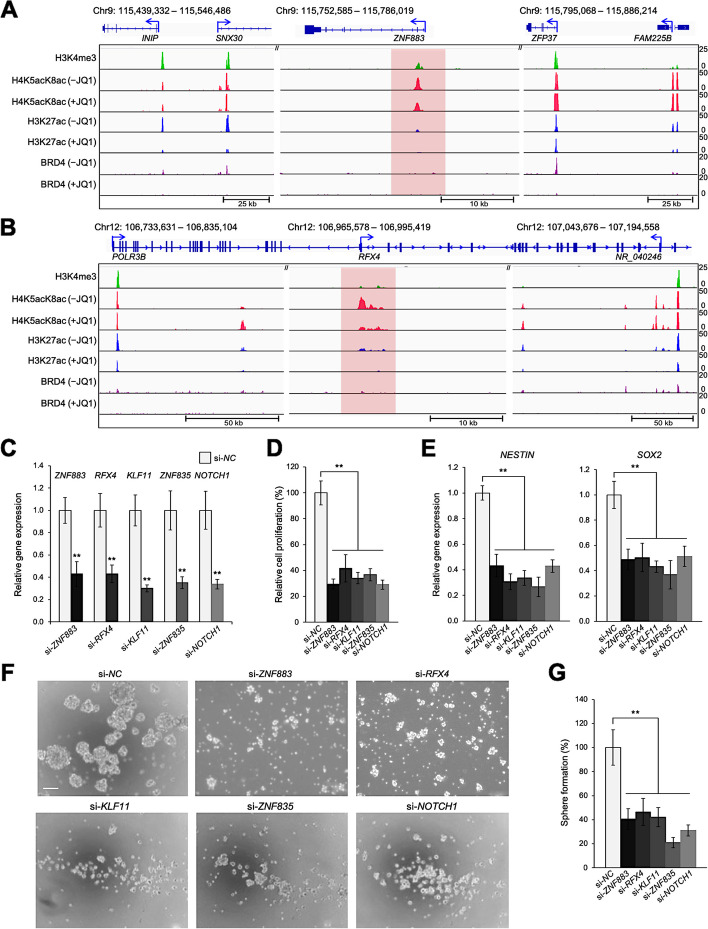
Fig. 4.
Disruption of glioblastoma stem-like properties by siRNA knockdown of genes with H4K5acK8ac-preferred promoters. A and B Comparative ChIP-seq occupancy tracks of H3K4me3, H4K5acK8ac, H3K27ac, and BRD4 at representative loci, in the presence or absence of JQ1. The promoter regions of ZNF883 (A) and RFX4 (B) were specifically enriched with H4K5acK8ac in 0316-GSC cells. The unique enrichment of H4K5acK8ac at promoters is highlighted in red. ChIP-seq reads were averaged from two biological replicates. C–E Disruption of gene expression by siRNA knockdown. C Efficiencies of siRNA knockdown of genes from Group 6 with H4K5acK8ac-preferred promoters and the GSC-specific control marker (NOTCH1) are compared with the negative control (si-NC) in 0316-GSC cells (n = 3). D Short-term proliferation assay of 0316-GSC cells subjected to siRNA knockdown. Cell proliferation rates at 7 days after siRNA knockdown of the selected genes are shown (n = 6). E Expression of stem cell marker genes, NESTIN and SOX2, following siRNA knockdown of the selected genes (n = 3). F and G Sphere formation assay following siRNA knockdown of the selected genes. F Phase-contrast images of 0316-GSC cells treated with target-specific siRNA. Images are representative of three independent experiments. Scale bar, 50 μm. G In vitro sphere formation efficiency of 0316-GSC cells treated with siRNA for 2 weeks (n = 3). C–E and G Data are means ± SEM **P < 0.01 (two-tailed Student’s t-test)