Figure 5. Cancer-associated mutations in HNRNPF/H genes.
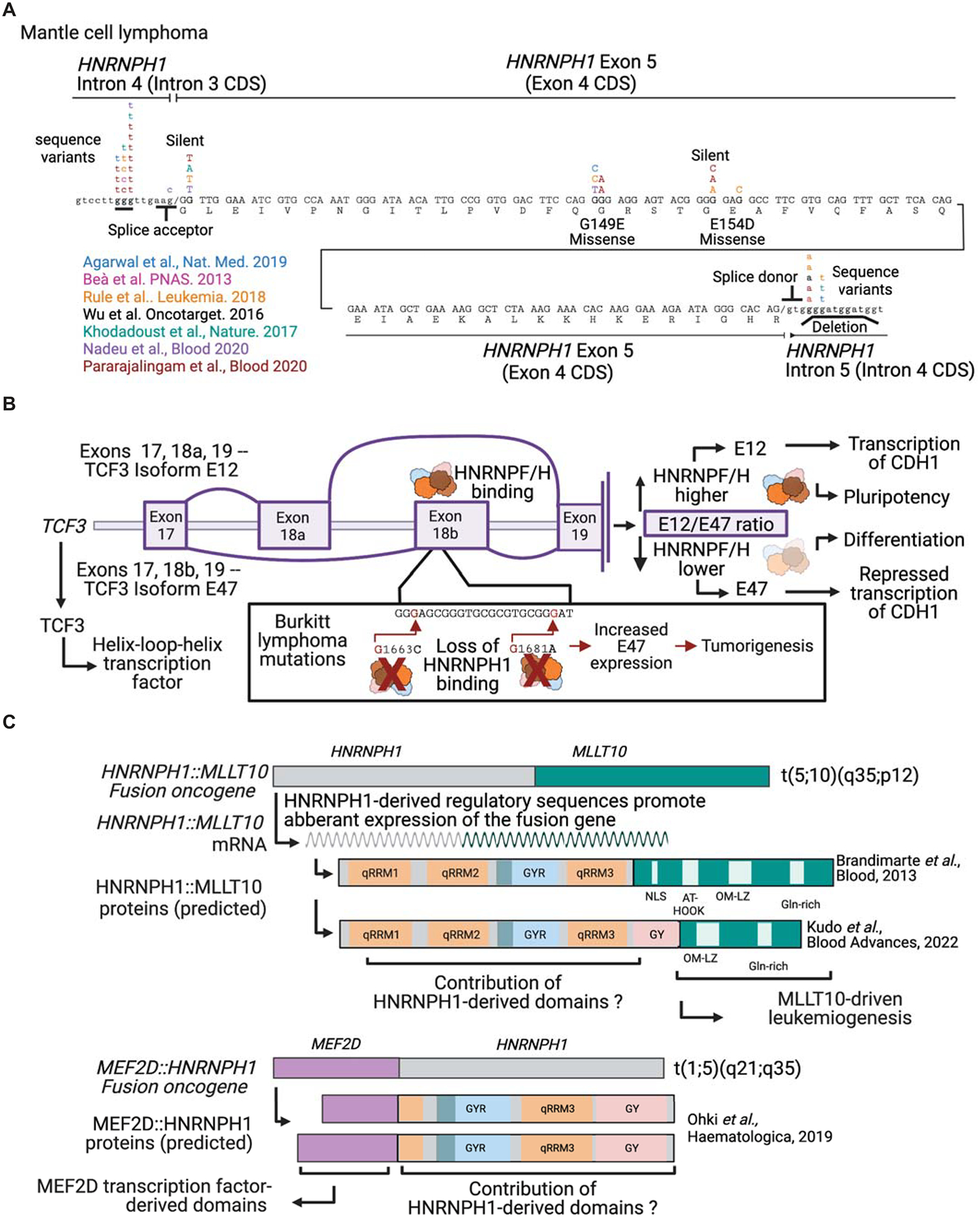
(A) A schematic of the position of mutations in the HNRNPH1 gene identified in mantle cell lymphomas reported by (Nadeu et al., 2020; Pararajalingam et al., 2020), with the latter study including reanalysis of the following studies (Agarwal et al., 2019; Beà et al., 2013; Khodadoust et al., 2017; Rule et al., 2018; Wu et al., 2016). (B) A schematic of the HNRNPF/H-dependent processing of TCF3 (transcription factor 3) transcript variants that express isoforms with different phenotypic outcomes and the effect of mutations that alter HNRNPF/H binding sites based on findings reported in Burkitt’s lymphoma(Yamazaki et al., 2020; Yamazaki et al., 2018; Yamazaki et al., 2019). (C) Schematics of MLLT10-HNRNPH1 fusion genes identified in pediatric T-acute lymphoblastic leukemia (Brandimarte et al., 2013) and pediatric acute myeloid leukemia (Kudo et al., 2022) and HNRNPH2-MEF2D identified in a case of pediatric B-cell precursor acute lymphoblastic leukemia (Ohki et al., 2019). MMT10 = MLLT10 histone lysine methyltransferase DOT1L cofactor; MEF2D = myocyte enhancer factor 2D. Figure panels created with BioRender.com.
