Figure 1.
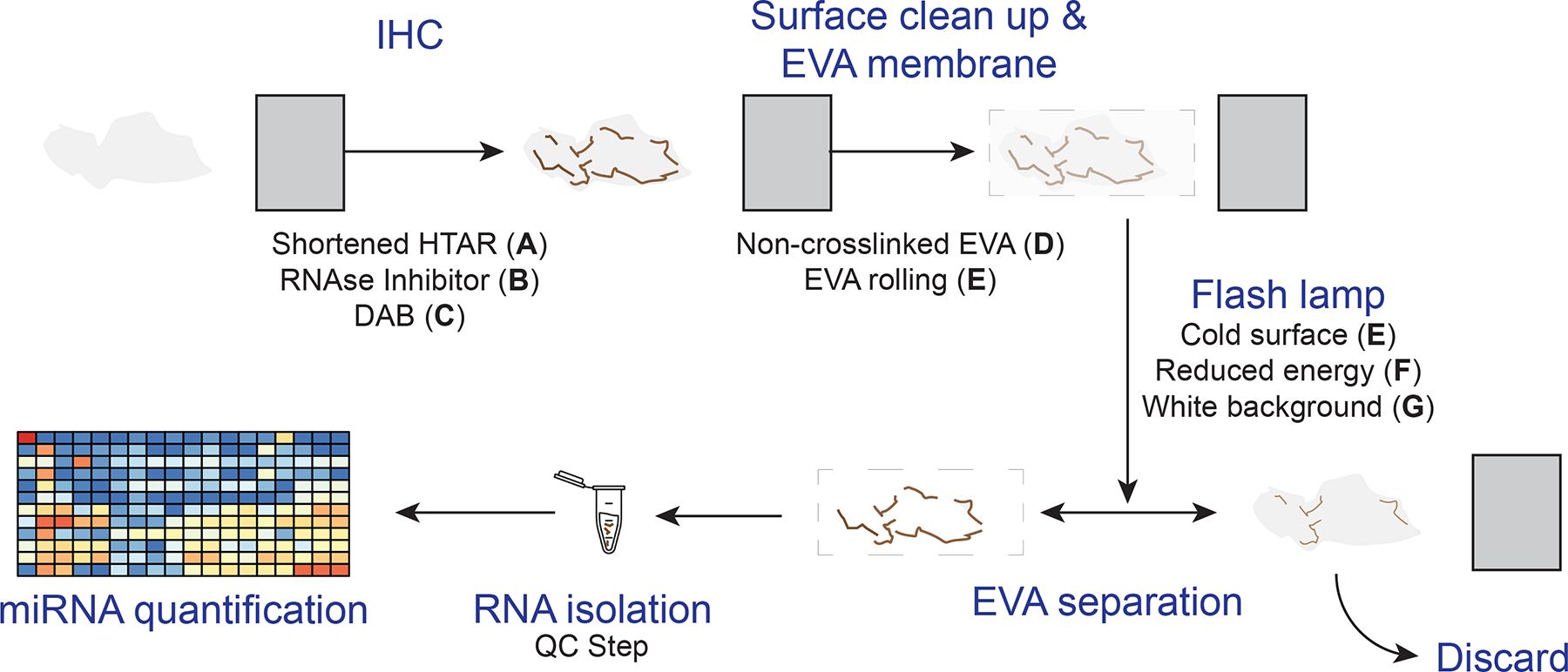
The general xMD method with improvements. A standard slide is generated from a formalin-fixed paraffin-embedded block on which IHC is performed for a cell type of interest. After IHC, the surface is cleared of ambient RNA, and an EVA membrane is tightly opposed. A flash lamp introduces light absorbed by pigmented cells that focally adhere to the EVA membrane. Then, the EVA membrane is dissolved to isolate RNA from the specific cell type for miRNA quantification. Improvements, described in this study in separate sections of the methods, were to shorten the HTAR time (A); add an RNAse inhibitor with the primary antibody (B); use DAB (C); create and use a noncrosslinked EVA membrane (D); and improve RNA isolation specificity by clearing the surface with EVA rolling (E) and using a cold surface (E); reducing the energy of the flash lamp (F), and using a white background for the flash (G). A quality control step before miRNA quantification was added. DAB. 3,3’-diaminobenzidine; EVA. ethylene vinyl acetate; HTAR, high-temperature antigen retrieval: IHC. immunohistochemistry; xMD. expression microdissection.
