Figure 1.
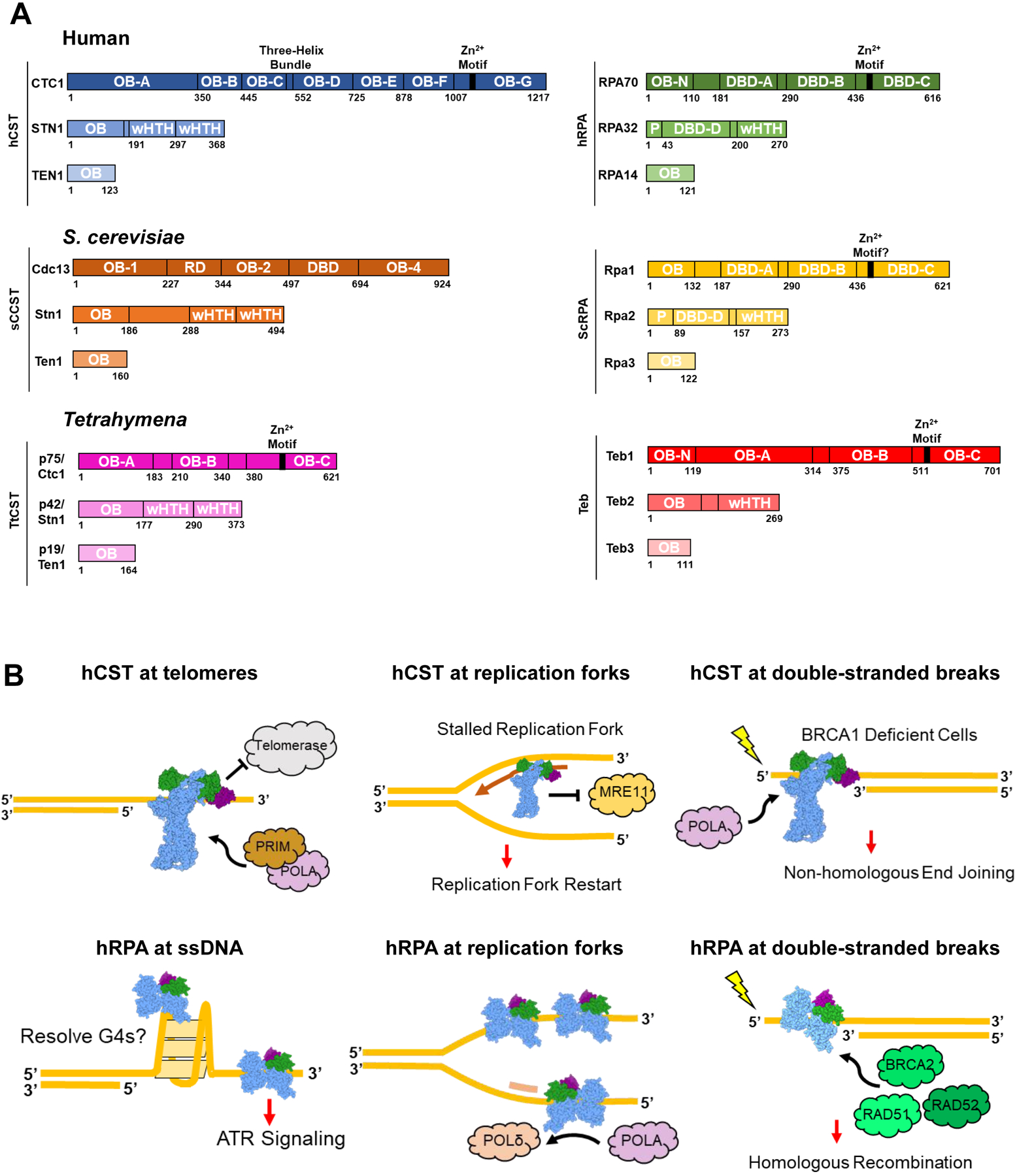
CST, RPA, and similar protein complexes across organisms. (A) Domain maps of the protein complexes explored in this review. hCST [37] in blue and hRPA [38,39] in green. ScCST [15,40,41] in orange and ScRPA [42] in yellow. TtCST [16] in pink and Teb [34,36] in red. OB = oligonucleotide/oligosaccharide binding fold; wHTH = winged helix-turn-helix; DBD = DNA-binding domain; P = phosphorylation domain; RD = recruitment domain. (B) Summary of hCST and hRPA functions at sites of ssDNA. Top left: hCST binds to the 3’ overhang of telomeres and terminates telomerase activity by blocking the binding site. hCST then facilitates the switch to C-strand fill-in by polymerase-α-primase (POLA, PRIM). Top middle: hCST aids in restarting stalled replication forks by recruiting polymerase-α-primase and blocking MRE11 from degrading nascent DNA. Top right: In BRCA1 deficient cells, hCST binds to single-stranded overhangs at double-stranded breaks which prevents end resection and facilitates fill-in by polymerase-α-primase. Ultimately this promotes non-homologous end joining as the DNA repair pathway. Bottom left: hRPA can bind and unfold G-quadruplexes as well as induce the ATR-repair pathway at long stretches of ssDNA. Bottom middle: hRPA stabilizes ssDNA at replication forks and facilitates the switch from polymerase-α-primase to polymerase-δ on the lagging strand. Bottom right: At sites of DNA damage, hRPA binds to the ssDNA and recruits repair proteins such as BRCA2, RAD51 and RAD52 which lead to the homologous recombination repair pathway.
