Figure 2.
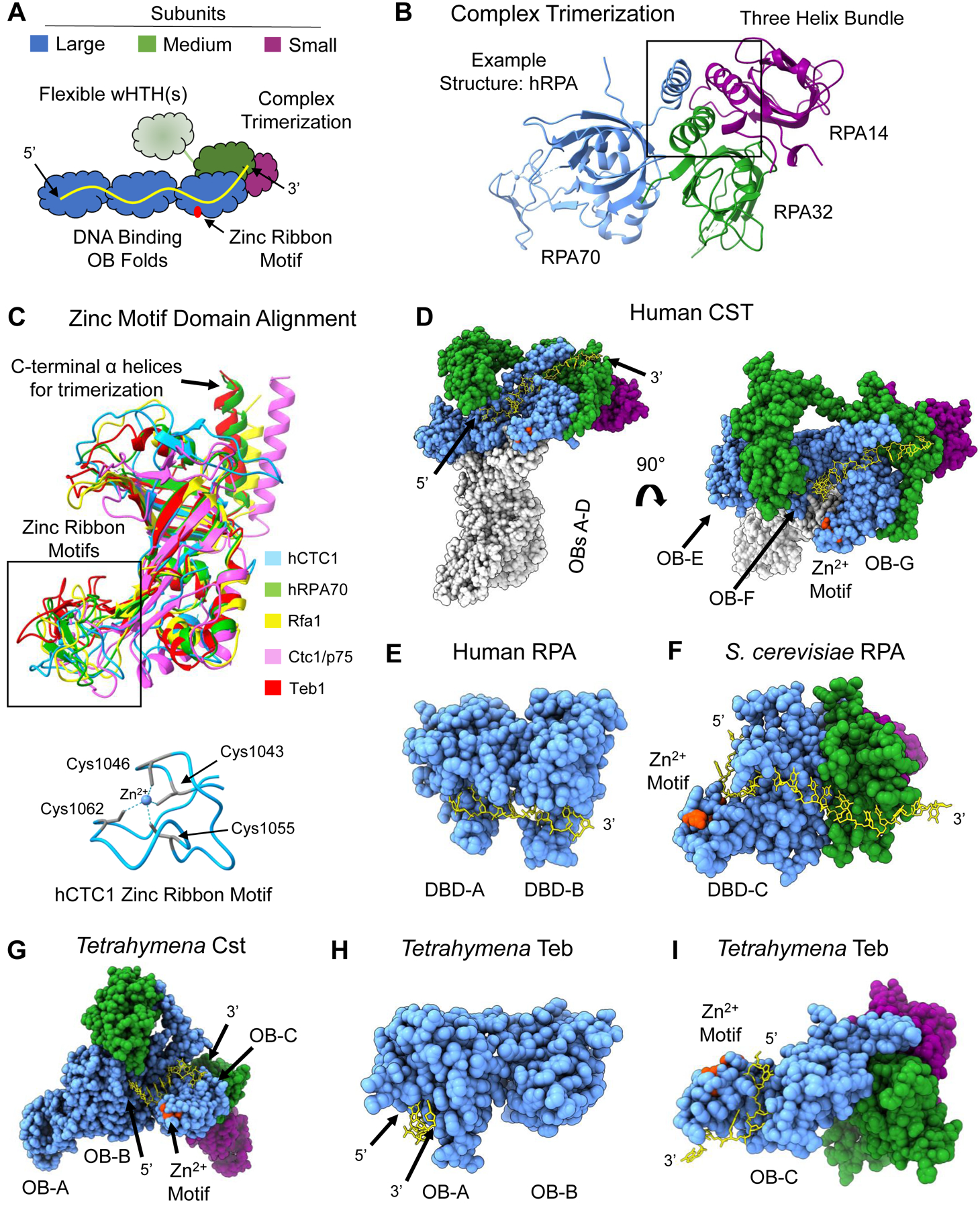
Shared structural features of RPA-like protein complexes. (A) Schematic of general architecture of RPA and RPA-like complexes. Three potential ssDNA-binding OB-folds are part of the largest subunit (blue) with a zinc ribbon motif (red) in the most C-terminal OB fold. The medium subunit (green) contains an OB fold that participates in complex trimerization with the large subunit and the smallest subunit (purple). The medium subunit contains either one or two wHTH domains that may undergo a conformational change via a flexible linker upon ssDNA (yellow) binding. (B) Three-helix bundle formation mediates trimerization between the three subunits. Example structure is from hRPA (PDB 1L1O). (C) Structure alignment of the zinc-ribbon motif containing OB folds of the five protein complexes in this figure. hCST in blue, hRPA in green, ScRPA in yellow, TtCst in pink and Teb in red. Below is the structure of the zinc ribbon motif of hCTC1 with the coordinating cysteine residues highlighted in teal with dashed lines representing interaction with the zinc ion in the center. An additional depiction of a zinc ribbon motif can be seen in RPA70 in panel B.(D) Structure of hCST bound to ssDNA (PDB 8D0B) from a face view (left) and a top view (right). The ssDNA spans OB-F and OB-G of CTC1 and the OB fold of STN1. It should be noted that this structure of hCST was solved as a co-complex with polymerase-a-primase, which is not shown here, and hCST may engage with ssDNA in a different manner without this protein-protein interaction. (E) Structure of hRPA with bound ssDNA resolved (PDB 1JMC). The ssDNA spans DBD-A and DBD-B of RPA70. The structure does not include DBD-C or either of the other subunits. (F) ScRPA bound to ssDNA (PDB 6I52). The ssDNA is bound to DBD-C of Rpa1 and the OB fold of Rpa2. (G) TtCST bound to ssDNA (PDB 7UY7). The ssDNA is bound to OB-B and OB-C. In this structure, the ssDNA does not contact Stn1 as seen in hCST. It should be noted that this structure of ttCST was solved in complex with polymerase-a-primase, which is not shown here. (H) Teb bound to ssDNA (PDB 3U58). The structure is only of OB-A and OB-B of Teb1 and depicts ssDNA in contact with OB-A alone. (I) Teb complex bound to ssDNA (PDB 6D6V). The structure only includes OB-C of Teb1 where the ssDNA is bound and the OB domain of Teb2 and Teb3. The zinc-ribbon motif is highlighted in red in D-I structures. Structure images and alignments generated in UCSF ChimeraX. Dashed lines in a structure refer to unresolved amino acids.
