Figure 3.
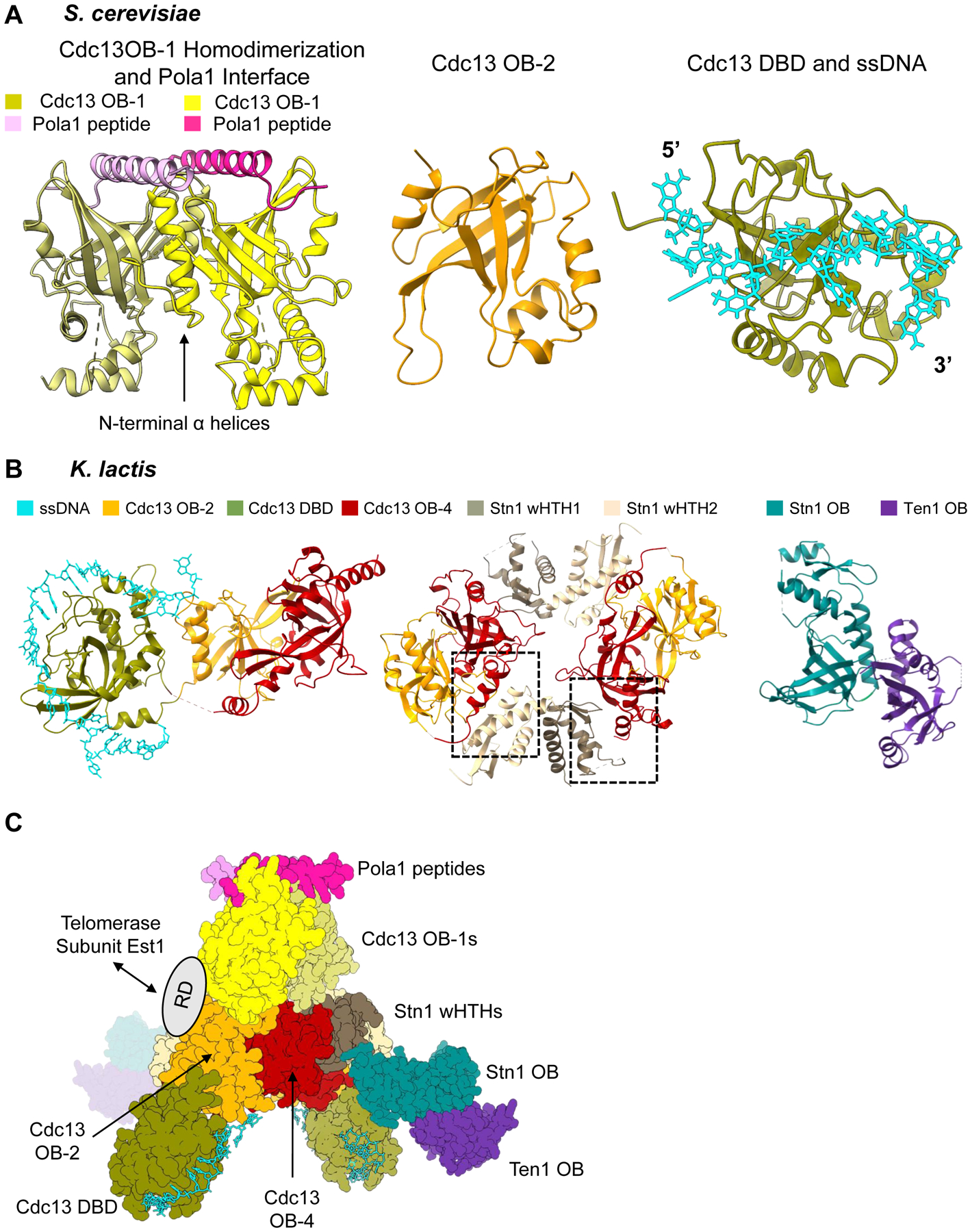
Piecing together the overall architecture of S. cerevisiae Cdc13-Stn1-Ten1. (A) Available structural information on S. cerevisiae Cdc13. Left: Dimerization of Cdc13 OB-1 and polymerase-α interaction site (PDB 3OIQ). N-terminal α-helices of OB-1 mediate the dimerization interface. The α-helix of a small polymerase-α peptide interacts with the Beta-barrel of the OB fold. Middle: OB-2 of Cdc13 (PDB 4HCE). Right: Cdc13 DBD bound to 11-nt ssDNA ligand 5’- GTGTGGGTGTG −3’ (PDB 1S40). (B) Structures of K. lactis Cdc13-Stn1-Ten1. Left: Cdc13 OB-2, DBD bound to 25-nt ssDNA ligand, and OB-4 (PDB 6LBR). The three domains form a horseshoe shape, allowing Cdc13 OB-2 and Cdc13 OB-4 to interact with each other. Middle: OB-2 and OB-4 of Cdc13 and the two winged-helix-turn-helix domains of Stn1 (PDB 6LBT). There are two unique hydrophobic interfaces (boxed) between Cdc13 OB-4 and each of the wHTHs that mediate a dimerization interaction. Right: Heterodimerization of OB fold domain of Stn1 and Ten1 (PDB 6LBU). (C) Overall architecture of Cdc13-Stn1-Ten1 proposed by Ge et al. 2020 using the solved structures of K. lactis and S. cerevisiae together. The ring dimerization interface of K. lactis Cdc13 OB-2 and OB-4 and Stn1 wHTHs is the center of the protein complex. The K. lactis DBD bound to ssDNA is then docked in based on its relationship with OB-2 and OB-4. The K. lactis Stn1 OB fold interacts with Stn1 wHTH-2 in the center ring, leaving Ten1 protruding from the center of the protein complex. The authors then used the S. cerevisiae dimerized OB-1 structure to show the possible interaction with the OB-2 domains in the ring. The polymerase-α interaction site with the OB-1 dimer is then accessible at one end of the protein complex. The flexible recruitment domain of Cdc13 would be accessible for interaction with Est1 on the outside of the dimerized protein complex. Structure images generated in UCSF ChimeraX. Dashed lines in a structure refer to unresolved amino acids.
