Figure 4.
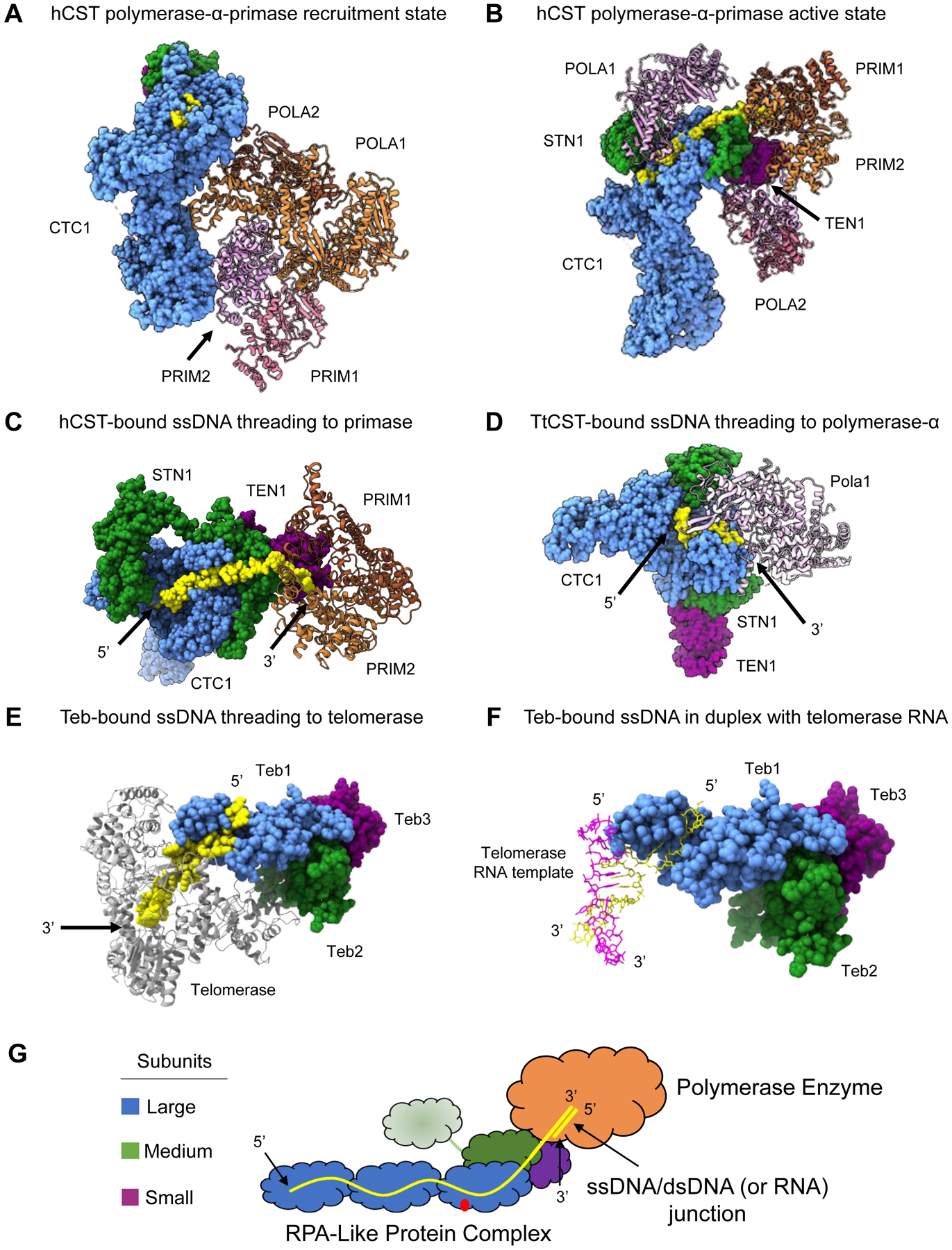
RPA-like protein complexes as processivity factors for polymerase enzymes. (A) hCST bound to polymerase-α-primase in the recruitment state. POLA1, POLA2 and PRIM2 make contacts with the OB folds of CTC1 (PDB 7U5C). (B) Polymerase-α-primase undergoes a sweeping conformational change into the active state in complex with hCST. PRIM1 and PRIM2 are stabilized by STN1 and TEN1 of hCST. POLA1 is bound to hCST in two distinct sites and POLA2 is in complex via interaction with POLA1 (PDB 8D0K). (C) hCST bound to polymerase-α-primase with telomeric ssDNA oriented to show the threading of ssDNA into the active site of primase (PDB 8D0K). This is the same complex as shown in Panel B, the POLA1 and POLA2 subunits are not shown to facilitate viewing the ssDNA (D) ttCST bound to the 5’ end telomeric ssDNA while the 3’ end is interacting with the active site of polymerase-α (PDB 7UY7). (E) ttTeb bound to telomeric ssDNA that is threaded into the active site of telomerase (PDB 6D6V). (F) ttTeb bound to telomeric ssDNA that forms a duplex with the telomerase RNA (PDB 6D6V). (G) Proposed model for how these RPA-like complexes act as a processivity factors to polymerase enzymes. The ssDNA is bound to the large subunit OB folds and may also bind the medium subunit, where the 3’ end is then threaded into the active site of the polymerase where duplex DNA may be generated by the enzyme. Structure images generated in UCSF ChimeraX. Dashed lines in a structure refer to unresolved amino acids.
