Abstract
Missense mutations in the DNA binding domain of p53 are characterized as structural or contact mutations based on their effect on the conformation of the protein. These mutations show gain-of-function activities, such as promoting increased metastatic incidence compared to p53 loss, often mediated by the interaction of mutant p53 with a set of transcription factors. These interactions are largely context specific. In order to understand the mechanisms by which p53 DNA binding domain mutations drive osteosarcoma progression, we created mouse models, in which either the p53 structural mutant p53R172H or the contact mutant p53R245W are expressed specifically in osteoblasts, yielding osteosarcoma tumor development. Survival significantly decreased and metastatic incidence increased in mice expressing p53 mutants compared to p53-null mice, suggesting gain of function. RNA-sequencing of primary osteosarcomas revealed vastly different gene expression profiles between tumors expressing the missense mutants and p53-null tumors. Further, p53R172H and p53R245W each regulated unique transcriptomes and pathways through interactions with a distinct repertoire of transcription factors. Validation assays showed that p53R245W, but not p53R172H, interacts with KLF15 to drive migration and invasion in osteosarcoma cell lines and promotes metastasis in allogeneic transplantation models. Additionally, analyses of p53R248W ChIP peaks showed enrichment of KLF15 motifs in human osteoblasts. Taken together, these data identify unique mechanisms of action of the structural and contact mutants of p53.
Significance
The p53 DNA binding domain contact mutant p53R245W, but not the structural mutant p53R172H, interacts with KLF15 to drive metastasis in somatic osteosarcoma, providing a potential vulnerability in tumors expressing p53R245W mutation.
Keywords: p53 hot spot mutations, KLF15, osteosarcoma, RNA-seq, promoter analysis, differential gain-of-function
Introduction
Tp53 is the most highly mutated gene in human cancers. Missense mutations in Tp53 are the most common alteration, with over 85% of mutations observed between codons 125 and 300, which encompass the DNA binding domain of the protein. This suggests that an altered form of the protein is more advantageous for tumor cells than deletion of the gene [1, 2]. Of the missense mutations in the DNA binding domain, 8 codons show a higher frequency of specific alterations and constitute about 28% of all p53 mutations [3]. These hotspot mutations can be classified according to their influence on the protein structure. Mutations at codons 248 and 273 are classified as contact mutations because of their occurrence at an arginine residue at the site of contact between protein and DNA. Mutations at residues R175H, G245S, R249S, and R252H severely impact the conformation of the DNA binding domain, and are classified as conformational or structural mutants [4, 5]. Both types of mutations abrogate the ability of the p53 protein to bind DNA, thereby causing loss of transcriptional activity. These hotspot mutants, however, retain the transactivation domain, and studies show that it is through this highly active transactivation domain, that mutant p53 alters the transcriptome of tumor cells [6].
Several studies show that cells with germline p53 missense mutations exhibit gain-of-function (GOF) activities by interacting with other proteins, many of which are transcription factors, and dysregulate gene expression to promote tumorigenesis and metastasis. Through this GOF, tumor cells expressing mutant p53 show a growth advantage, and increased resistance to therapeutic drugs in vitro as compared to p53-null cells [7-9]. In breast cancer cells, p53R172H interacts with SMAD to mediate TGF-β induced metastasis [10], and with Vitamin D receptor (VDR) to convert vitamin D into an anti-apoptotic agent [11]. The p53R273H mutant in breast cancer cells binds SREBP transcription factors to increase sterol biosynthesis via the mevalonate pathway [12] and can also interact with SP1 to promote HIV replication [13].
In vivo, germline p53R172H/+ and p53R270H/+ mutations (corresponding to human hotspot mutations Tp53R175H and Tp53R273H, respectively) develop tumors with increased metastasis as compared to p53+/− mice [14, 15]. In germline osteosarcoma, p53R172H binds to ETS2 to regulate a phospholipase Pla2g16 that results in increased metastasis [16]. Thus, multiple mechanisms contribute to mutant p53 GOF activities in context-specific manner.
Osteosarcoma is an aggressive type of malignant bone tumor that shows rapid tumor development and early pulmonary metastasis in humans. This five-year survival rate after chemotherapy drops from 70% to approximately 25% with metastatic disease [17]. The most common genes recurrently mutated in osteosarcoma are the tumor suppressors p53 and Rb which are considered to be drivers of this disease [18]. Mutations in the transcription factor p53 in particular, correlate with high levels of genomic instability, thereby playing a significant role in initiation and/or propagation of osteosarcomas [19]. Treatment options for osteosarcoma have plateaued for over twenty years, and it is vital to understand the contributions of different p53 mutations in driving osteosarcoma.
Genetically engineered mice with somatic mutations in p53 have shed light on different p53 mutants and their effects on tumorigenesis and metastasis. In osteosarcomas, Rb and p53 loss specifically in osteoblasts led to early-onset osteosarcomas with complete penetrance [20].
Tumors of mesenchymal origin, like osteosarcomas, were studied by Pourebrahim et al. to explore the effects of p53R172H on osteosarcomagenesis and metastasis [21]. In this study, mice expressing a single allele of mutant p53 in osteoblasts develop osteosarcomas with 100% penetrance by 13 months of age, and show an increased metastatic incidence as compared to mice lacking p53 in osteoblasts. The increased metastatic incidence is completely reversed with genetic abrogation of the ETS2 transcription factor [21]. Taken together, different p53 mutants are known to have different effects on cells in vitro as well as in vivo. It is of vital significance to understand the GOF activities of mutant p53 at the molecular level as they may present novel therapeutic vulnerabilities in mutant p53 driven tumors.
Here we present a comparative analysis of the differences in abilities of structural and contact mutants of p53 to drive osteosarcoma in vivo and in vitro. We describe novel genetically engineered somatic mouse models of osteosarcoma expressing either the contact mutant p53R245W or the structural mutant p53R172H in osteoblasts. We show that not only are the transcriptomes of tumors with a p53 missense mutation different from those of p53-null tumors, the transcriptomes, as well as the pathways affected by the gene expression changes, for p53R245W expressing tumors, are unique compared to p53R172H expressing tumors. Further, we identify a distinct repertoire of transcription factors that the respective p53 missense mutants interact with to regulate the unique transcriptomes. Finally, we identify and validate KLF15 as a novel interacting partner of p53R245W, but not p53R172H in murine somatic osteosarcoma. Downregulation of p53R245W or Klf15 decreases migration, invasion, and metastasis in different assays. Our findings are recapitulated in human osteoblasts and a human osteosarcoma cell line, providing clinical relevance.
Materials and Methods
Mice
All animal studies were approved by, and were performed in compliance with Animal Care and Use Committee (IACUC) at University of Texas, MD Anderson Cancer Center. The wm (wild-type to mutant) alleles were generated and characterized as described previously [21, 22]. mTmG, and TdTomato transgenic mice (RRID:IMSR_JAX:007909) as well as transgenic Osx-Cre mice (RRID:IMSR_JAX:006361) were purchased from Jackson Laboratories. Mice with both p53 alleles floxed out (RRID:IMSR_JAX:008462) have been previously described [23]. Animals were monitored daily for tumor formation by palpation, as well as for malocclusion. Tumors, along with other relevant tissues were excised and processed for histology as well as generation of cell lines, and the remaining tissue was snap frozen in liquid nitrogen, before storage at −80°C for further genomic, transcriptomic, gene expression, and proteomic analyses.
Genotyping
Genotyping of all mice was performed by Polymerase Chain Reaction, using primers surrounding the 5’ lox-P sites for all p53 alleles. The primers used for genotyping the p53 alleles, as well as the Rosa26 locus, are listed in Supplementary table 1.
Tail-vein injections in athymic nude mice
NU/J athymic nude mice (Strain #:002019 RRID:IMSR_JAX:002019) were requested from Jackson Labs. All animal studies were approved by, and were performed in compliance with Animal Care and Use Committee (IACUC) at University of Texas, MD Anderson Cancer Center. 500,000 cells were suspended in sterile ice-cold PBS (1X) and injected into the tail-vein of mice. Mice were monitored daily for tumor formation, and hyperpnea. Lungs were excised and stored in 10% formalin for analysis.
Mouse osteosarcoma cell lines and tissue processing
Tissue preps were performed as described previously [21]. Micro-CT scans were performed by the Small Animal Imaging Facility at MD Anderson Cancer Center using approved protocols. Cell lines were established from murine primary osteosarcomas upon excision of tumor. A small piece of the tumor was cut and washed twice in a 10cm tissue culture plate with sterile 1X PBS. It was then chopped finely using a blade in 3ml Trypsin/EDTA solution at room temperature. The crushed tissue was allowed to solvate in Trypsin/EDTA for 5-7 minutes at 37°C. 10ml DMEM with 10% FBS, and 1% Penicillin/Streptomycin solution was added to Trypsin/EDTA to stop the reaction. The plate was then placed in the incubator at 37°C with 5% CO2 levels. After 24 hours, the plate was checked for cells established on the surface, and the media was refreshed.
RNA isolation
The primary osteosarcomas stored at −80°C were used to isolate RNA. Approximately 100ug of the frozen tissue was crushed into a fine powder on dry ice using a CellCrusher. The powder was dissolved in Trizol reagent before being processed for RNA isolation using the Direct-sol RNA Microprep kit (R2060, Zymo Research) as per recommendations from the kit manufacturer. The RNA quality assessment and quantification were performed on the Bio-Analyzer (RRID:SCR_019715).
RNA-seq analysis and pipeline
RNA-seq libraries were generated by the Advanced Technology Genomics Core (ATGC) at MD Anderson Cancer Center. Quality of paired end reads was verified using fastqc (RRID:SCR_014583). Adapter contamination and low-quality reads were removed using Trim Galore! (RRID:SCR_011847). Reads were aligned to the mm10/GRCm38 mouse genome using STAR aligner (RRID:SCR_004463). Read quantification was performed using HTSeq (RRID:SCR_005514). DESeq2 (RRID:SCR_015687) was used to identify differentially expressed genes. Differentially expressed genes were defined by a False Discovery Rate (FDR) < 0.05 and Log Fold Change < −2 or > 2. R language for statistical computing (RRID:SCR_001905) was used for DESeq2 analysis. Ingenuity Pathway Analysis (RRID:SCR_008653) was used to perform pathway analysis using differentially expressed genes. Ensembl Biomart (RRID:SCR_004727) was used to obtain 10kb DNA sequences of the upstream flanks of the differentially upregulated genes. MEME-Suite (RRID:SCR_001783) was used to perform promoter analysis to identify potential interacting partners of mutant p53. MEME-Suite (v5.5.1) and related tools were used to identify enriched motifs [24].
Western blots (WB), co-immunoprecipitation assays, quantitative reverse transcriptase PCR (qRT-PCR), and RNA interference
WB and qRT-PCR analyses were done as described previously [25]. Protein A/G agarose beads (Santa Cruz Biotechnology Cat# sc-2003, RRID:AB_10201400) were used for co-immunoprecipitation assays, for which protocols have been described previously [26]. The antibodies for WB analysis were p53, 1:1000 (Leica Biosystems Cat# NCL-L-p53-CM5p, RRID:AB_2895247), KLF15, 1:1000 (Santa Cruz Biotechnology Cat# sc-271675, RRID:AB_10710379), Vinculin, 1:10,000 (Sigma-Aldrich Cat# V9131, RRID:AB_477629) and β-actin (Sigma-Aldrich Cat# A2066, RRID:AB_476693). Primers used for qRT-PCR analysis are described in Supplementary Table 1. siRNAs directed against p53 as well as universal controls for all RNA interference experiments in this study have been described previously [25]. siRNAs directed against Klf15 (SASI_Mm01_00138081, and SASI_Mm01_00138083, Sigma Millipore) were applied using protocol similar to si-p53. Transfection protocols to generate stable knockdown of p53 has been described previously [27]. Virus targeting Klf15 for stable knockdown was requested from the Functional Genomics Core at MD Anderson Cancer Center and transfected using standard protocols.
Migration assays
10,000 - 25,000 cells were incubated in 300ul serum-free DMEM (with 1% Penicillin/Streptomycin) in BioCoat transwell (354578, Corning) for 18 hours at 37°C. The underside of the transwell was submerged in 600ul of DMEM with 10% Fetal Bovine Serum. After incubation, the underside of the transwell was fixed and stained with 6% glutaraldehyde, and 0.5% crystal violet for 30 minutes at room temperature. The transwells were then washed 5 times with 1X PBS. The cells on the inner surface of the transwell were removed by gently swiping with a cotton q-tip. Cell migration was determined using Image J software (RRID:SCR_003070) by ‘Percent Area Migrated’ using a Nikon Plate Reader to image the entire transwell.
Invasion assays
50ul Matrigel (354230, Corning) was thawed at 4°C for 4 hours, before plating in BioCoat transwell (354578, Corning) and allowing to solidify at 37C for 30 minutes. 10,000 - 25,000 cells were layered on the Matrigel in 300ul serum-free DMEM (with 1% Penicillin/Streptomycin). The underside of the transwell was submerged in 600ul of DMEM with 10% Fetal Bovine Serum. After incubation, the underside of the transwell was fixed and stained with 6% glutaraldehyde, and 0.5% crystal violet for 30 minutes at room temperature. The transwells were then washed 5 times with 1X PBS. The cells on the inner surface of the transwell were removed by gently swiping with a cotton q-tip. Cell migration was determined using Image J software (RRID:SCR_003070) by ‘Percent Area Invaded’ using a Nikon Plate Reader to image the entire transwell.
ATAC-sequencing and analysis
100,000 cells were submitted in biological triplicates for ATAC-seq. Library preparation for ATAC-sequencing was performed by the Epigenomics Profiling core at MD Anderson Cancer Center. Protocols for library preparation, sequencing and analysis have been described previously [28].
Chromatin immunoprecipitation (ChIP)-qRT-PCR
300,000-500,000 cells were used for crosslinking with 1% formaldehyde with protease inhibitors for 10 minutes at room temperature before quenching the reaction with 125mM Glycine for 5 minutes. The cells were then Dounce homogenized and resuspended in 750ul of ice-cold immunoprecipitation buffer (100 mM Tris, pH 8.6, 0.3% sodium dodecyl sulfate [SDS], 1.7% Triton X-100, 5 mM ethylenediaminetetraacetic acid [EDTA]). These were then sonicated to an average length of 200-500bp (confirmed by agarose gel electrophoresis) by pulsing 40 times for 30 seconds followed by 30 seconds of rest and 50% amplitude on a sonicator. Approximately 1mg protein was used for the assay which was conducted in biological triplicates. Antibodies used were KLF15, 1:1000 (Santa Cruz Biotechnology Cat# sc-271675, RRID:AB_10710379), H3 (Abcam, Cat# ab1791 RRID:AB_302613) and Normal Rabbit IgG (Cell Signaling Technology Cat# 2729, RRID:AB_1031062). qRT-PCR was performed as described previously [25]. Primer sets are listed in Supplementary table 1.
Generation of p53R248W/+ (p53R248W) and wild-type (WT) human embryonic stem cells (hESCs)
H1 (WA01) hESCs (WA01) were purchased from WiCell. hESCs were cultured on Matrigel-coated plates (Corning) in StemMACS iPS-Brew XF medium (Miltenyi Biotec) at 37°C in a humidified 5% CO2 incubator. Cells were passaged using StemMACS™ Passaging Solution XF (Miltenyi Biotec) when reaching 85% confluence. hESC clones with p53 point mutations were generated using TALENs as previously described [29]. Upstream and downstream TALEN target sites used to introduce edits leading to monoallelic p53R248W mutation of Tp53 are 5’-tccaggtcaggagccact-3’ and 5’-ggcctgctgtgcccca-3’ at intron 7. Donor vectors were constructed using site-directed mutagenesis to generate p53R248W (CGG > TGG). Corresponding WT hESC clones were generated using WT donor vectors.
Human ESC culture and in vitro differentiation to derive human osteoblasts
Mesenchymal stem cells (MSCs) were derived from hESCs using the methods previously described [30]. Briefly, hESC clones were first cultured in StemMACS iPSC-Brew XF medium supplemented with 10 uM TGFβ inhibitor SB-431542 (Sigma-Aldrich) on Matrigel-coated plates at 37°C and 7.5% CO2. The majority of cells demonstrate a spindle-shaped morphology at 20 days, when they were passaged to gelatin-coated plates and cultured in MSC differentiation medium (KnockOut DMEM/F12 (Gibco) supplemented with 10% KnockOut serum replacement, β-mercaptoethanol, non-essential amino acids, penicillin-streptomycin, glutamine, 20 ng/ml EGF, and 20 ng/ml bFGF) in the presence of SB-431542 for another 20 days. Cells were checked periodically for MSC surface markers CD105 and CD166, and when the majority of cells were positive they were designated as passage 0 MSCs. MSCs were cultured in aMEM (Corning) containing 10% FBS at 37°C and 5% CO2. For osteogenic differentiation, MSCs were seeded in 6-well plates at a density of 20,000 cells per well and cultured in osteogenic differentiation medium (aMEM supplemented with 10% FBS, 10 mM β-glycerol phosphate, 200 uM ascorbic acid, and 0.1 uM dexamethasone) for 24 days [31, 32].
ChIP-seq for p53 chromatin occupancy in human osteoblasts
ChIP-sequencing was performed as previously described with some modifications [30]. Osteoblasts were fixed in 1% formaldehyde for 10 minutes at room temperature and then quenched by 125mM glycine. Fixed cells were collected and sonicated in ChIP lysis buffer (50mM HEPES-KOH pH 7.5, 140mM NaCl, 1mM EDTA, 0.1% Na-Deoxycholate, 0.1% SDS, 1% Triton X-100, protease inhibitor cocktail) on ice. The lysate supernatant was incubated overnight at 4°C with anti-p53 antibody (Santa Cruz Biotechnology, Cat# sc-126, RRID:AB_628082). The mixture of cell lysates and antibodies was then diluted with the same amount of ChIP dilution buffer (50mM HEPES-KOH pH 7.5, 140mM NaCl, 1mM EDTA, 1% Triton X-100) and incubated with magnetic protein G dynabeads (Thermo Fisher Scientific, Cat# 10004D) for 3 hours at 4°C. The immunoprecipitate on the beads was washed with ChIP high salt buffer (20mM Tris-HCl pH 8.0, 500mM NaCl, 2mM EDTA, 0.1% SDS, 1% Triton X-100) and treated with RNase A and proteinase K. DNA was eluted in TE buffer containing 1% SDS and purified by QIAquick PCR purification kit (QIAGEN). Purified DNA was used for library preparation and sequencing at the BGI Genomics. NGS reads were aligned to hg19 using bowtie2, peaks were called using MACS2, and annotated for adjacent genes using HOMER. Peaks locations were compared using bedtools, genomic DNA sequences for regions spanned by peaks were downloaded from UCSC genome browser, and motif enrichment was performed using MEME-Suite.
Over-expression of human mutant of p53 hotspots in Saos-2 cells
Saos-2 cells expressing p53R175H, and p53R248Q have been described previously [16]. Wild-type Saos-2 cells were authenticated using short tandem repeat (STR) profiling by the Cytogenetics and Cell Authentication Core at MD Anderson Cancer Center in January 2023. NSK026 CMV-TO-p53R248W (FLP-IN) was a gift from Xiaojing Gao (Addgene plasmid # 191168; http://n2t.net/addgene:191168; RRID:Addgene_191168) [33], and transfected in Saos-2 cells to express p53R248W, using standard procedures.
Immunohistochemistry
Anti-p53 antibody, 1:1000 (CM5, Leica), was used for staining overnight, and hematoxylin was used as a counterstain for the primary tumors. Quantification of cells with stable p53 was done using Image J software, on five non-overlapping fields at 10X objective magnification using light microscopy. Scoring was performed based on percentage of p53-positive cells divided by the total number of cells in the field. Tumors with <25% p53-positive cells were scored 1, 25-50% were scored 2, 50-75% were scored 3, and >75% were scored 4.
Statistical Analysis
Student t-tests, Fisher’s Exact tests, and ANOVAs were performed using Prism 9 (RRID:SCR_002798). Differences were considered significant at a value of p < 0.05.
Data availability
The data generated in this study are publicly available in National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) at GSE231852. All other raw data are available upon request from the corresponding author.
Results
p53 mutants in a mouse model of osteosarcoma show a metastatic phenotype in a p53-stability dependent manner
To study the role of p53 mutants p53R172H and p53R245W in osteosarcoma development, we generated mice with these mutations in osteoblasts using the conditional p53wm (wild-type to mutant) alleles that allow for expression of the mutant p53 after Cre-mediated recombination (Supplementary figure 1A). Expression of TdTomato from the Rosa26 locus provides a marker for cells that have undergone Cre-mediated recombination. We established the following cohort: p53fl/+Osx-CreTgRosa26TdT/+ (hereafter referred to as Op53+/−, n = 13), p53wmR172H/+Osx-CreTgRosa26TdT/+ (hereafter referred to as Op53R172H/+, n = 13), and p53wmR245W/+Osx-CreTgRosa26TdT/+ (hereafter referred to as Op53R245W/+, n = 15) (Supplementary Table 2). This cohort allowed us to examine the effects of p53 dosage, and loss of heterozygosity (LOH) in osteosarcomagenesis and survival. In addition, we set up the following cohort: p53fl/flOsx-CreTgRosa26TdT/+ (hereafter referred to as Op53−/−, n = 40), p53wmR172H/flOsx-CreTgRosa26TdT/+ (hereafter referred to as Op53R172H/−, n = 18), and p53wmR245W/flOsx-CreTgRosa26TdT/+ (hereafter referred to as Op53R245W/−, n = 27) (Supplementary Table 2) to study the role of mutant p53 in absence of WT p53. We obtained primary osteosarcomas at multiple sites such as the pelvis, spine, jaw, chest and long bones of arms and legs (Figure 1A, Supplementary figure 1B). Most tumors of the spine could only be detected at a microscopic level, as these tended to cause paralysis in mice, necessitating euthanasia.
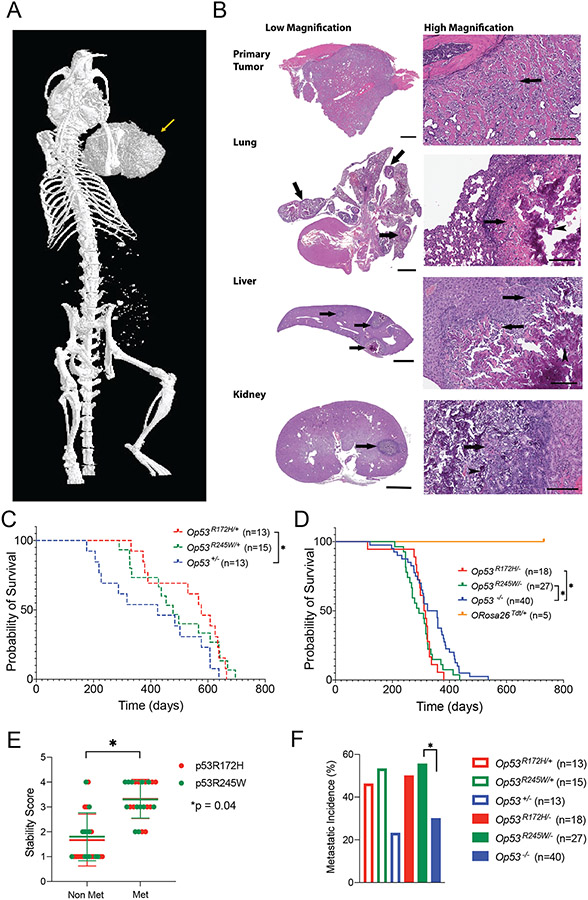
Figure 1:
Osteosarcomas with different p53 missense mutations metastasize with high frequency. A: Osteosarcoma with p53R245W/+ mutation, in the long bone of the left arm (arrow) as observed using micro-CT imaging of whole mouse. B: Representative photomicrographs (10-20X for low magnification, 200X for high magnification) of an Op53R245W/+ mouse that developed osteosarcoma. Discrete tumor nodules (arrows) compress and invade non-mineralized native structures in lung, liver, and kidney (low magnification). Under high magnification, neoplastic cells show expansile growth pattern with centrally located mineralized, basophilic osteoid regions (arrowheads), surrounded by non-mineralized eosinophilic osteoid regions (arrows); scale bars: low magnification primary tumor, 900 microns; lung, liver, and kidney, 2000 microns; high magnification, 200 microns. C and D: Survival curves for different genotypes that yield osteosarcomas. E: Stability scores based on immunohistochemistry analysis for primary osteosarcomas expressing either p53R172H (N = 24) or p53R245W (N = 24) mutations. F: Metastatic incidence observed in mice with different genotypes. *p < 0.05.
A metastatic phenotype was observed in this cohort; metastatic lesions were observed in lungs, liver and kidneys by hematoxylin and eosin staining (Figure 1B). All mice with metastatic osteosarcomas had metastases in lungs. Incidentally, lungs are the primary organ for metastasis in human osteosarcomas as well [34].
Histopathological analyses of the primary tumors and metastatic lesions revealed features typically observed in human osteosarcomas. In the metastatic lesions observed in the mouse models, the discrete tumor nodules were compressed and minimally invaded the adjacent native structures observed under low magnification (Figure 1B). Under high magnification, neoplastic cells were observed to produce variable amounts of extracellular matrix (osteoid) that formed thin borders around neoplastic cells or broad irregular trabeculae between groups of neoplastic cells. Consistent with the typical expansile growth pattern of metastatic nodules of osteosarcoma, the more mature, centrally located osteoid were mineralized, and therefore basophilic (arrowheads), while regions of less mature, non-mineralized (eosinophilic) osteoid (arrows) were usually located more peripherally. These data suggested that the metastatic lesions in the soft tissue organs were skeletal in origin.
Mice with Op53R172H/+ outlived Op53+/− significantly (p = 0.04) (Figure 1C). The median survival for Op53+/− mice was 424 days while the median survival for Op53R172H/+ and Op53R245W/+ mice was 577 and 478 days, respectively. There were no differences in survival for Op53R245W/+ and Op53+/− mice. Survival is a parameter reflective of tumor growth rate, but is not indicative of end-stage metastatic disease especially in mice. This likely explains the extended survival observed in Op53R172H/+. In mice expressing only mutant p53, survival decreased as expected, and Op53R172H/− mice (median survival 313.5 days; p = 0.02 by log-rank test) and Op53R245W/− mice (median survival 293 days; p = 0.02 by log-rank test) died earlier than Op53−/− mice (median survival 340 days). This suggested an enhanced tumorigenic potential for the p53 missense mutants compared to p53 deletion. Mice that expressed only TdTomato (Osx-CreTgRosa26TdT), with WT p53 after Cre-mediated recombination did not exhibit tumor development during the analysis period (Figure 1D). Additionally, we did not observe differences in survival for mice that expressed TdTomato versus mice that did not express TdTomato (Supplementary figure 1C). This suggested that TdTomato was not tumorigenic by itself in our tumor model.
To examine the effects of LOH of WT p53, we examined 9 and 8 primary osteosarcomas from Op53R172H/+, and Op53R245W/+ groups respectively, by Sanger sequencing. Based on the criteria used for calling LOH described previously [35], we identified 22% (2/9) and 38% (3/8) of Op53R172H/+ and Op53R245W/+ tumors respectively, retained the p53 WT allele. 78% (7/9) and 62% (5/8) of Op53R172H/+ and Op53R245W/+ tumors respectively, showed loss of the WT p53 allele (Supplementary Figure 1D). Statistical significance between retention or loss of the WT p53 allele was not observed, suggesting that the mutants were capable of driving tumorigenesis irrespective of the presence of WT p53 allele.
Previous studies have shown that mutant p53 can be stable within the tumor cell, and stability of mutant p53 is a pre-requisite for its GOF potential [36]. To check the stability of mutant p53, we performed immunohistochemistry staining on 24 primary tumors, each expressing either p53R172H and p53R245W over p53 WT or floxed alleles. We designated a score of 1 to the primary osteosarcomas that showed stable p53 in less than 25% of the cells in five non-overlapping fields; a score of 2 to tumors that showed between 25-50% cells with stable p53, a score of 3 to tumors that showed 50-75% cells with stable p53, and a score of 4 to tumors with more than 75% cells with stable p53. Based on criteria for scoring the stability of p53 in primary tumors, we did not observe differences between the stability of p53R172H and p53R245W. However, we observed that metastatic primary osteosarcomas had a higher stability score compared to non-metastatic primary osteosarcomas (p = 0.04) (Figure 1E).
Based on the number of mice analyzed for metastases in Supplementary Table 3, we calculated the metastatic incidence in our cohort (Figure 1F, Supplementary Table 3). About 50% of the Op53mut/+ mice showed pulmonary metastases, trending higher than Op53+/− mice (23%). In the absence of a WT p53 allele, we saw a similar trend in metastatic incidence. Mice from Op53R245W/− group (55.6%) had a significantly higher metastatic incidence compared to Op53−/− group (30%) (p = 0.04, by Fisher’s Exact Test). Mice from Op53R172H/− group also trended with higher metastases (50%) but was not significant compared to p53-null mice. The trends observed in the metastatic incidence combined with the strong correlation of p53 stability with metastatic tumors, and survival data, suggested that both p53R172H and p53R245W had a greater tumorigenic potential as compared to p53 loss alone.
Primary osteosarcomas expressing p53R172H or p53R245W have different gene expression profiles compared to p53-null tumors
In order to understand the mechanisms driving enhanced tumorigenic and metastatic potential of p53 missense mutants, we performed RNA-sequencing on primary osteosarcomas and compared the gene expression data to p53-null primary osteosarcomas. We previously published data on 16 primary osteosarcomas (8 Op53−/−, 8 Op53R172H/−) which show p53R172H metastatic gain of function is mediated via ETS2 [21]. We treated these samples as batch 1. An additional 18 primary osteosarcoma samples (2 Op53−/−, 5 Op53R172H/−, and 11 Op53R245W/−) were sequenced and labeled batch 2. (Differences in survival and metastatic incidence were not observed between the old and new cohorts – Supplementary Table 2, Supplementary Figure 2A, 2B). Six samples per group from the new cohort (Figure 1, Supplementary Table 2) were added as a third batch. This brought our total number of samples to 16 Op53−/−, 19 Op53R172H/−, and 17 Op53R245W/− primary osteosarcomas. We took every precaution to not introduce batch effects in the RNA-sequencing analysis. To assess for batch effects, tumors of each genotype were plotted in principal component analysis plots (Supplementary Figure 2C, 2D, and 2E). Plots demonstrate the majority of samples cluster regardless of batch with no predilection for outliers in each genotype to come from any particular batch, suggesting an absence of batch effects.
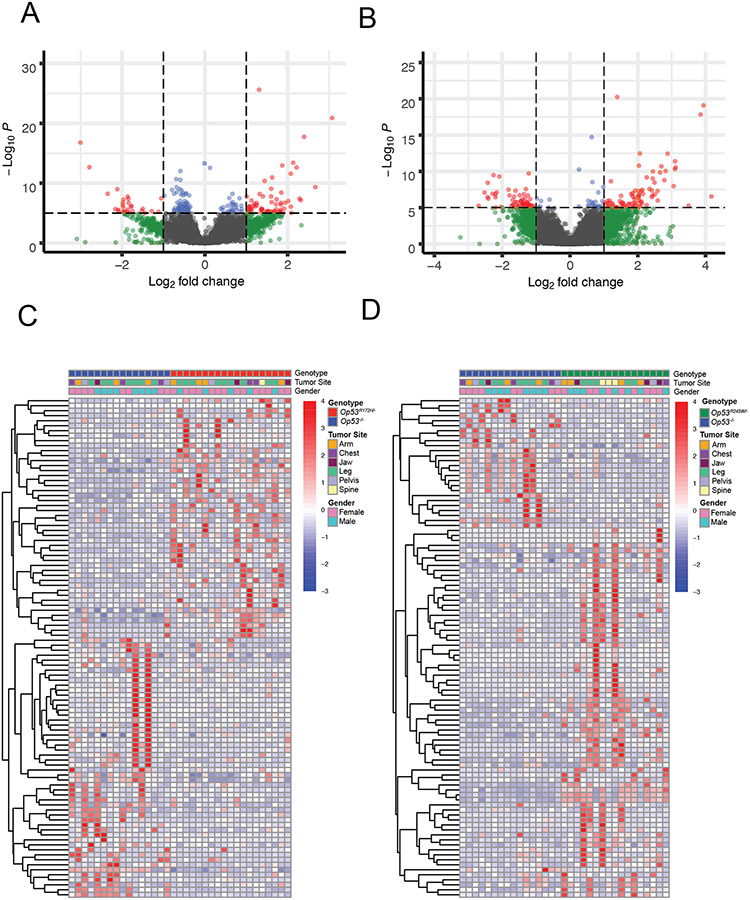
Neither p53R172H nor p53R245W binds DNA directly. Instead these missense mutant proteins bind other proteins many of which are transcription factors and regulate their transcription which contributes to tumorigenesis and/or metastasis. Furthermore, the repertoire of transcription factors that these missense mutants bind is broad, mutant-specific, tissue-specific in most cases, and is largely understudied [1, 2]. To identify mechanisms enabling the gain of function potential of p53 missense mutants, we compared the gene expression data from tumors expressing the p53 missense mutants to those tumors lacking p53 expression. The volcano plots for p53R172H and p53R245W show the distribution of genes in the two groups (Figure 2A and 2B). At a false discovery rate (FDR) of 5%, a significant number of genes were observed to be differentially expressed (differentially expressed genes; DEGs) – 1564 DEGs for Op53R172H/− and 1679 DEGs for Op53R245W/− at log2 fold change (LFC) 0; 594 DEGs for Op53R172H/− and 999 DEGs for Op53R245W/− at LFC < −1 or > 1. In order to reduce noise and increase specificity, we applied strict significance criteria of FDR < 5%, and LFC greater than 2 for upregulated genes or less than −2 for downregulated genes. Using these criteria, we identified 255 DEGs (156 upregulated; 99 downregulated) in Op53R172H/− and 409 DEGs (275 upregulated; 134 downregulated) in Op53R245W/− when compared to tumors with Op53−/−. These data suggested significant differences in gene expression between tumors that expressed p53 missense mutants and p53-null tumors. To visualize differences, we ranked the top 100 DEGs by absolute LFC for Op53R172H/− and Op53R245W/− compared to Op53−/− and plotted them as heatmaps (Figure 2C and 2D).
Figure 2:
Transcriptomic analyses of primary murine osteosarcomas show that transcriptomes of those expressing p53 missense mutations differ significantly from those that do not express p53. A and B: Volcano plots showing distribution of gene expression differences in Op53R172H/− and Op53R245W/− osteosarcomas, respectively. C and D: Heatmaps showing differential expression of top 100 genes ranked by absolute LFC in Op53R172H/− v. Op53−/− and Op53R245W/− v. Op53−/− osteosarcomas, respectively.
p53R172H and p53R245W regulate unique transcriptomes, dysregulate different pathways, and potentially bind to distinct sets of transcription factors
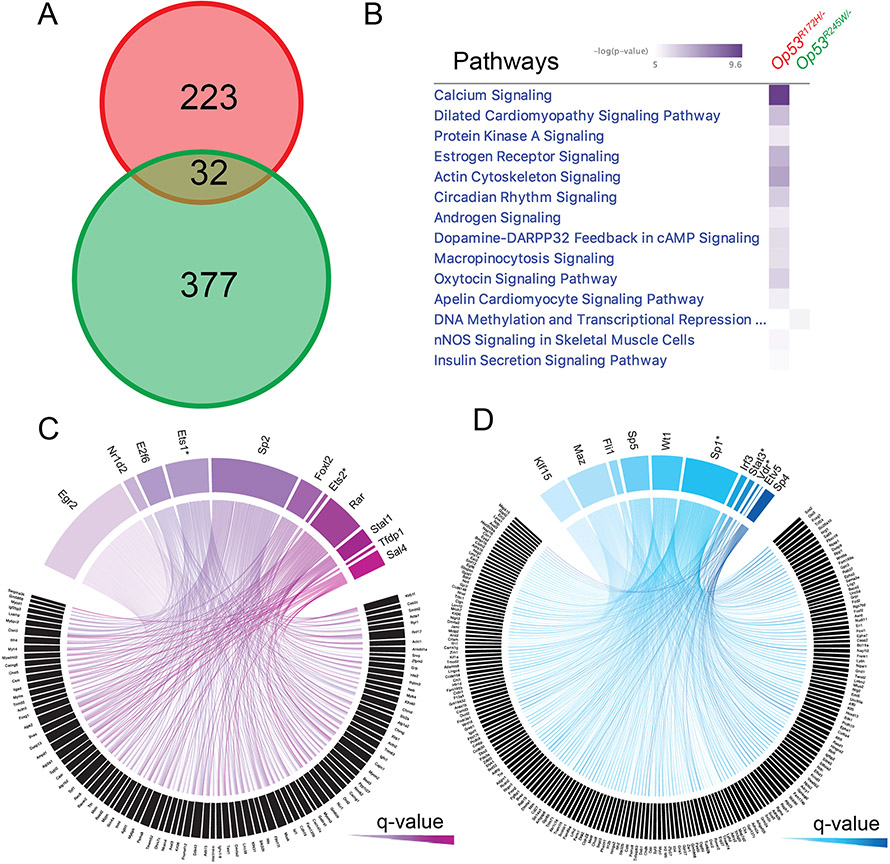
In order to understand the mechanisms driving the differential expression of these genes, and the pathways that these genes affect, we first examined the common genes that were differentially expressed in both groups. Surprisingly, we identified only 32 DEGs that were common to both groups at an FDR of 5%, and LFC < −2 or > 2 (Figure 3A). Ingenuity Pathway Analysis (IPA) using these 32 genes found no significant pathways were identified at a -log(P-value) of 5.0 (Supplementary Figure 2F). This suggested that the overlapping genes do not contribute to differences in pathway activation in our tumors, causing us to focus on mutant-specific DEGs. To visualize the contrast between Op53R172H/− and Op53R245W/− after comparing to the Op53−/− controls, we plotted the top 187 genes in the union of the top 100 for each group (13 DEGs that were present in both lists) (Supplementary Figure 3A). We next performed IPA using two separate inputs: all DEGs from Op53R172H/− and all DEGs from Op53R245W/−. The top signaling pathways with a -log(P-value) > 5.0 that were dysregulated in Op53R172H/− were not dysregulated in Op53R245W/− (Figure 3B). This suggested that p53R172H and p53R245W regulate different transcriptomes and dysregulate different pathways to exert their tumorigenic potential.
Figure 3:
Transcriptomes regulated by p53R172H and p53R245W mutants are different. A: Venn diagram showing minimal intersection of the differentially expressed genes in Op53R172H/− (red) and Op53R245W/− (green) osteosarcomas, p = 1.00E+00. B: Dysregulated pathways in Op53R245W/− compared to Op53R172H/−, based on differing -log(P-value). C and D: Circos plots depicting the differentially upregulated genes in Op53R172H/− and Op53R245W/− osteosarcomas, and the list of transcription factors, arranged by significance, binding to enriched motifs in a 10kb region upstream of the transcription start sites of differentially upregulated genes.
Previous studies have shown that mutant p53 proteins bind different transcription factors in order to execute their gain of function and upregulate genes that aid in tumorigenesis and metastasis [1, 2]. Since these mechanisms are largely context-dependent and our expression profiles show more difference than similarity between Op53R172H/− and Op53R245W/−, we performed a promoter analysis on the upregulated genes in each p53 missense mutant to identify the transcription factors that regulate the differentially upregulated genes observed in these murine somatic osteosarcomas. Briefly, we analyzed a 10kb region upstream of the transcription start site (TSS) of all upregulated genes for enriched motifs. We then identified the set of transcription factors that bound to these enriched motifs. With a q-value cutoff of 0.01, we identified 11 unique transcription factors that had enriched motifs in the 10kb regions upstream of TSSs of the differentially upregulated genes in each Op53R172H/− and Op53R245W/− (Figure 3C and 3D; Table 1). EGR2 was the top transcription factor with a q-value of 8.88E-11 in the Op53R172H/−, while KLF15, with a q-value of 7.38E-06 was the top transcription factor in the Op53R245W/−. Further, the upregulated genes had enriched motifs for multiple transcription factors with no overlap between the mutants. Two transcription factors, ETS2 and VDR were previously identified to interact with mutant p53, thus increasing confidence in our analysis [11, 16]. Further, we were able to identify novel transcription factors like EGR2, and NR1D2 that may interact with p53R172H, and MAZ and FLI1, that may interact with p53R245W (Table 1). Taken together, these data suggest that p53R172H and p53R245W can potentially bind and co-opt distinct transcription factors, to regulate unique transcriptomes and dysregulate different pathways during osteosarcomagenesis and metastasis in our mouse model.
Table 1:
Transcription factors (TFs) with enriched motifs in p53 mutant osteosarcomas. Colored TFs (purple) were identified in analysis excluding outliers (Supplementary Figure 3B). Asterisks represent TFs known to interact with mutant p53 in published studies.
| TFs with enriched motifs in p53R172H tumors |
q-value | TFs with enriched motifs in p53R245W tumors |
q-value |
|---|---|---|---|
| EGR2 | 8.88E-11 | KLF15 | 7.38E-06 |
| NR1D2 | 2.84E-09 | MAZ | 7.74E-06 |
| E2F6 | 6.48E-07 | FLI1 | 1.01E-05 |
| ETS1* | 2.68E-06 | SP5 | 0.00019677 |
| SP2 | 5.76E-05 | WT1 | 0.00019677 |
| FOXL2 | 5.76E-05 | SPP1* | 0.00066578 |
| ETS2* | 0.0005007 | IRF3 | 0.00246145 |
| RARA | 0.00091714 | STAT3* | 0.00246145 |
| STAT1 | 0.00120136 | VDR* | 0.00295375 |
| TFDP1 | 0.00458336 | ETV5 | 0.00365936 |
| SALL4 | 0.00530985 | SP4 | 0.00592509 |
In our RNA-sequencing analysis, we identified 3 outliers in each group (Supplementary figure 2C, 2D, and 2E). Outliers were from various batches with the majority of samples overlapping irrespective of batch. Excluding the outliers from our analysis, we identified 7 and 9 of 11 transcription factors (from Table 1) with a q-value cut-off of 0.01, for p53R172H and p53R245W tumors, respectively. A list of top transcription factors with a q-value cut-off of 0.01, when outliers are excluded, is shown in Supplementary Figure 3B. The concordance observed suggested the inclusion of outliers in our analysis did not skew the results of our promoter analysis.
Validation of gene expression and chromatin differences between p53 mutants
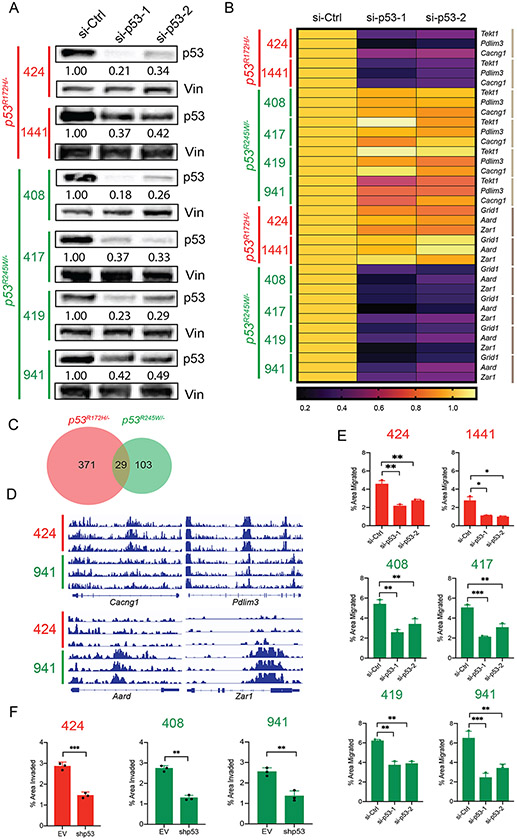
We established mouse osteosarcoma cell lines from primary mouse osteosarcomas in an effort to develop a validation system. Cell lines were established from tumors with Op53−/− (# 561, 1606), Op53R172H/− (# 424, 1441) and Op53R245W/− (# 408, 417, 419, 941) genotypes thereby allowing us to recapitulate the comparison groups. The cell lines expressing either of the missense mutants of p53 showed a stable p53 protein, which could be depleted using 2 independent si-RNAs targeting mutant p53 (Figure 4A and Supplementary figure 4A).
Figure 4:
Knockdown of mutant p53 in murine osteosarcoma cell lines results in reduction of migration potential. A: Immunoblots of p53 protein levels after treatment with two independent siRNAs against p53 in murine osteosarcoma cell lines expressing either p53R172H (424, 1441 – in red) or p53R245W (408, 417, 419, 941 – in green). B: qRT-PCR analysis of gene targets of p53R172H, Tekt1, Pdlim3, Cacng1 (denoted by light brown lines on right), and gene targets of p53R245W Grid1, Aard, Zar1 (denoted by dark brown lines on right) upon knockdown of respective p53 mutants; Gapdh served as control. C: Overlap between the number of genes in the open chromatin regions for cells expressing p53R172H (red) and p53R245W (green). D: Loci depicting the peaks corresponding to open chromatin regions for gene targets of p53R172H, Cacng1, and Pdlim3 (top panel), and for gene targets of p53R245W, Aard, and Zar1 (bottom panel) E: Murine osteosarcoma cell lines expressing p53R172H (red) or p53R245W (green) and their migration potential upon knockdown of p53. F: Murine osteosarcoma cell lines expressing p53R172H (red) or p53R245W (green) and their invasion potential upon knockdown of p53. Significance calculated by student’s t-test from at least 3 biological replicates; *p < 0.05, **p < 0.01, ***p < 0.005.
Further, we tested the effect of mutant p53 knockdown on gene expression as well. Three genes that were amongst the top DEGs in Op53R172H/− and Op53R245W/− were tested for gene expression changes upon mutant p53 knockdown. Tekt1, Pdlim3, and Cacng1 were amongst top differentially upregulated genes in Op53R172H/− while Grid1, Aard, and Zar1, were amongst the top differentially upregulated genes in Op53R245W/−. As seen in Figure 4B (and Supplementary Figure 4B), upon mutant p53 knockdown, the expression of these genes was significantly downregulated in the respective groups. All three genes, Tekt1, Pdlim3, and Cacng1 showed downregulation in gene expression upon mutant p53 knockdown in cell lines expressing p53R172H, but not in cell lines expressing p53R245W. Additionally, Grid1, Aard, and Zar1 were downregulated in cell lines expressing p53R245W, but not p53R172H. This analysis in osteosarcoma tumor cell lines, supported our in vivo data showing that different p53 missense mutants regulated different genes.
Our preliminary gene expression analysis showed that the respective p53 mutants regulated different gene sets in murine osteosarcoma cell lines. To further interrogate the differences in gene expression observed between the p53 mutant tumors, we performed ATAC-seq using cells expressing either the p53R172H mutant (# 424) or the p53R245W mutant (# 941) and identified open chromatin regions in the two cell lines when compared to a p53-null murine osteosarcoma cell line (# 561). Using a strict significance criterion of FDR < 1% and LFC > 3.0, we identified 400 genes in the open chromatin region in cells expressing p53R172H, 132 genes with open chromatin in cells that expressed p53R245W, and a small overlap of only 29 genes between the two groups (Figure 4C). We also visualized individual loci for genes of interest from our gene expression analysis (Figure 4D). Gene targets of p53R172H, Cacng1 and Pdlim3 show clear peaks corresponding to the open chromatin regions in the p53R172H-expressing cell line # 424, but not in the p53R245W-expressing cell line # 941 (Figure 4D, top panel). Likewise, we observed peaks corresponding to open chromatin regions for gene targets of p53R245W (Aard, Zar1) in cells expressing this mutant (# 941), but not in cells expressing p53R172H (# 424) (Figure 4D, lower panel). Next, we interrogated the transcription factors that were found upstream of the transcription start sites of genes identified in the open chromatin regions for cells expressing either p53R172H or p53R245W. Only 2 transcription factors (EGR2 and SP2) were nominated by open chromatin analysis of both mutants (q-value cutoff of 0.01; Table 2), a result that is consistent our RNA-seq finding that the two p53 mutants regulate unique sets of genes through different mechanisms. Additionally, we identified several transcription factors that overlapped with our RNA-seq findings. (Table 2).
Table 2:
Transcription factors (TFs) with enriched motifs in genes found in the open chromatin regions of cell lines expressing p53 mutants. Asterisks denote overlap with Table 1.
| TFs with enriched motifs in p53R172H cells |
q-value | TFs with enriched motifs in p53R245W cells |
q-value |
|---|---|---|---|
| EGR2* | 3.34E-06 | SP5* | 7.73E-04 |
| IRF3 | 8.45E-03 | ETV5* | 8.23E-03 |
| STAT1* | 2.41E-03 | KLF15* | 1.56E-01 |
| GATA6 | 7.49E-01 | FOXL2 | 0.00096321 |
| RARA* | 1.86E-01 | SP2 | 0.00021432 |
| SP2* | 0.0008432 | EGR2 | 0.0052388 |
| ETS1* | 0.0053966 | MAZ* | 0.0025436 |
| TFDP1* | 0.0035211 |
Mutant p53 knockdown shows reduction in migration and invasion potential
Since mutant p53 gain of function has been implicated in metastasis, we tested if mutant p53 knockdown had any phenotypic effect on our cell lines. We performed migration assays on cells with knockdown of mutant p53 and observed a significant reduction in migration potential of the cells with decreased levels of mutant p53 (Figure 4E). Of note, while all cell lines with knocked-down mutant p53 showed decreased migration, the inherent migration potential differed between the cell lines. Nonetheless, these data suggested that mutant p53 contributed to increased migration in these cell lines. Additionally, we also generated stable knockdown of mutant p53 in cells expressing either p53R172H (# 424), or p53R245W (# 408, 941) with 60-80% attenuation of protein expression (Supplementary figure 4C) to examine invasion. Invasion assays were performed using Matrigel and these stable knockdowns showed a significantly reduced capacity to invade through Matrigel upon mutant p53 depletion (Figure 4F), supporting our hypothesis that mutant p53 contributes to their invasive phenotype.
KLF15 binds to p53R245W, but not p53R172H, to mediate its ability to drive metastasis
p53R172H executes its metastatic gain of function mediated partially via transcription factor ETS2 in murine somatic osteosarcomas [21]. However, the mechanism by which p53R245W exerts its gain of function remains to be elucidated. From our promoter analysis of the RNA-sequencing data from primary murine osteosarcomas, we identified KLF15 as a transcription factor with the most significantly enriched motif in the 10kb region upstream of TSSs of upregulated genes in the p53R245W group but not in the p53R172H group (Figure 3C and 3D). This suggested that p53R245W could potentially bind to KLF15 to regulate the differentially upregulated genes identified in our experiment.
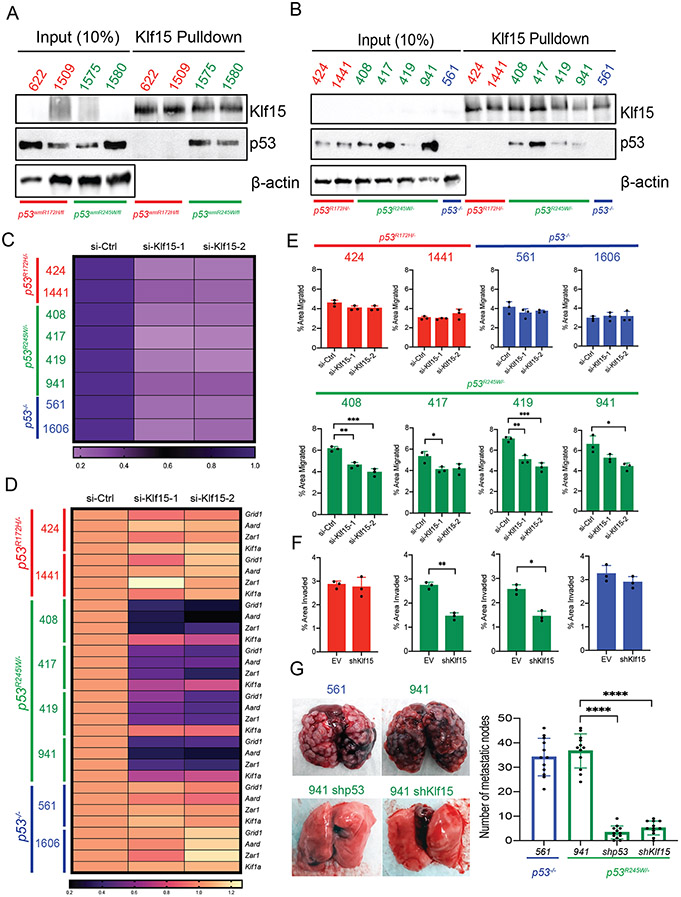
To investigate the role of KLF15 in murine somatic osteosarcoma, and to determine if p53R245W interacts with KLF15, we performed co-immunoprecipitation assays on primary tumors expressing p53R172H (tumor numbers 622 and 1509) and p53R245W (tumor numbers 1575 and 1580). Our previous results indicated that primary osteosarcomas that metastasized, also showed stable p53 protein (Figure 1E). As such, we selected tumors with a high p53 stability score for the co-immunoprecipitation assays. To examine interactions, we pulled down KLF15 protein, and probed the immunoblots with anti-p53 antibody. As seen in Figure 5A (and Supplementary Figure 5A), p53R245W, but not p53R172H bound to KLF15. We also performed the same co-immunoprecipitation assays in osteosarcoma cell lines with stable mutant p53. For this assay, we also included a p53-null murine osteosarcoma cell line (# 561) as a negative control. As seen in Figure 5B (and Supplementary Figure 5B), p53 protein was detected in cell lines expressing p53R245W (# 408, 417, 419, 941) but not p53R172H (# 424, 1441) or p53-null control cell line (# 561) after immunoprecipitation with a KLF15 antibody. KLF15 was expressed at very low levels in murine primary osteosarcomas as well as cell lines.
Figure 5:
KLF15 selectively binds to p53R245W but not p53R172H to execute mutant p53 gain of function. A: Co-immunoprecipitation assays in primary murine osteosarcomas expressing stable p53R172H (622, 1509 – in red) or p53R245W (1575, 1580 – in green) mutants. Pulldown with KLF15 antibody (top) and probe with p53 antibody (middle); β-actin served as loading control (bottom). B: Co-immunoprecipitation assay in murine osteosarcoma cell lines with stable p53R172H (424, 1441 – in red) or p53R245W (408, 417, 419, 941 – in green) mutants or p53-null cell line (561 – in blue). Pulldown with KLF15 antibody (top) and probed with p53 antibody (middle); β-actin served as loading control (bottom). C: Expression of Klf15 upon treatment with two independent siRNAs against Klf15 as quantified by qRT-PCR in murine OS cell lines expressing p53R172H (in red), p53R245W (in green) mutants or no p53 (561, 1606 – in blue); Gapdh used as control. D: Gene expression analysis by qRT-PCR in murine osteosarcoma cell lines for transcriptional targets of KLF15 and p53R245W – Grid1, Aard, Zar1. Kif1a is a gene target of p53R245W but is not a transcriptional target of KLF15 and was used as a negative control; Gapdh served as control. E: Effects of Klf15 knockdown on migration potential of murine osteosarcoma cell lines. F: Effects of Klf15 knockdown on invasion potential on murine osteosarcoma cell lines expressing p53R172H (red), p53R245W (green) or null for p53 (blue). G: Mouse lungs with metastatic nodes after tail-vein injections of p53-null cells (blue) or cells expressing p53R245W (green), with a stable knockdown of p53R245W, or stable knockdown of Klf15 (green) quantified on the right. Significance calculated by student’s t-test from experiments using at least 3 biological replicates; *p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.001.
Further, we wanted to test if Klf15 knockdown had any effect on its target genes. The significantly upregulated gene targets of p53R245W, Grid1, Aard, and Zar1, described in our previous set of experiments, were also found to have KLF15 binding motifs in the 10kb region upstream of their TSSs. Therefore, these were ideal candidates to test if KLF15 actually regulated the expression of these genes. Since we could not easily detect KLF15 protein levels under basal conditions in our mouse osteosarcoma cell lines, we checked efficacy of Klf15 knockdown using two independent si-RNAs by qRT-PCR. Klf15 mRNA levels showed a significant reduction when treated with both Klf15 siRNAs, as compared to scrambled controls (Figure 5C, Supplementary figure 5C) in all our cell lines. To test the effect of Klf15 knockdown on target gene expression, we performed qRT-PCR, and introduced another gene, Kif1a, that did not have KLF15 binding motif in the 10kb region upstream of its TSS, as a negative control. As seen in Figure 5D (and supplementary figure 5D), expression of target genes Grid1, Aard, and Zar1 was significantly downregulated in si-Klf15 groups in cell lines that expressed p53R245W (# 408, 417, 419, 941), but not in cell lines that expressed p53R172H (# 424, 1441) or p53-null cell lines (# 561, 1606). The expression of Kif1a that was used as a negative control, remained relatively unchanged. Thus, KLF15 specifically regulated the Grid1, Aard, and Zar1 genes that were significantly upregulated by p53R245W. Taken together, these data showed that p53R245W interacted with KLF15 and regulated expression of its target genes.
Next, we wanted to test if Klf15 knockdown had an effect on the metastatic potential of the cells. We hypothesized that in murine osteosarcoma cell lines, p53R245W, but not p53R172H would exert its gain of function effects via KLF15. As expected, Klf15 knockdown, showed a significant decrease in migration potential of cells expressing p53R245W but not p53R172H or p53-null cell lines (Figure 5E). Taken together, these data suggest that p53R245W, but not p53R172H binds to KLF15 to execute its gain of function, which in our cell lines, is their ability to migrate.
To test the effects of KLF15 on invasion, we generated stable knockdown of Klf15 in cells expressing the respective p53 mutants (# 424 expressing p53R172H; # 408, 941 expressing p53R245W) as well as in p53-null cells (# 561). In these cell lines, we achieved 50-60% knockdown efficiency (Supplementary figure 6A) and performed Matrigel invasion assays. We observed a reduction in invasion potential of cells that expressed the p53R245W mutant (# 408, 941), but not in cells that expressed p53R172H (# 424) or p53-null cells (# 561) (Figure 5F). This further supported our hypothesis that KLF15 interacted with p53R245W to mediate the ability of the cells to migrate/invade. Finally, we also performed tail-vein injections in athymic nude mice to examine the role of p53R245W as well as KLF15 on metastasis. We used the p53R245W expressing murine osteosarcoma cell line # 941 as a positive control. We had also used this cell line to generate stable knockdowns of p53R245W (Supplementary figure 4C) and Klf15 (Supplementary Figure 6A). Additionally, we also included a p53-null cell line (# 561) as an additional control. We observed a significant reduction in pulmonary metastasis in mice that were injected with cells that had knockdown of either p53R245W or Klf15 as compared to cells expressing the mutant, or p53-null cells (Figure 5G).
KLF15 binds the Zar1 promoter and p53R248W ChIP peaks are enriched for KLF15
To investigate the binding of KLF15 at the promoters of its target genes, we performed ChIP-qRT-PCR on Zar1, a known target of p53R245W as well as KLF15. MEME analysis identified two binding sites for KLF15 in the 10kb region upstream of the transcription start site of Zar1. To examine binding, we knocked down Klf15 using 2 independent siRNAs (Supplementary Figure 6B) in cells expressing p53R172H (# 424), p53R245W (# 408, 941), and p53-null cells (# 561). In p53R245W mutant cell lines, we observed a significant reduction in signal at the distal binding site of Zar1 upon knockdown of Klf15 (Supplementary figure 6C). Klf15 knockdown did not show differences in signal at the proximal binding site, nor did we observe any differences after Klf15 knockdown in cells expressing p53R172H or p53-null cells. These data demonstrate the localization of KLF15 to the Zar1 promoter in a manner that is specific to cells expressing p53R248W.
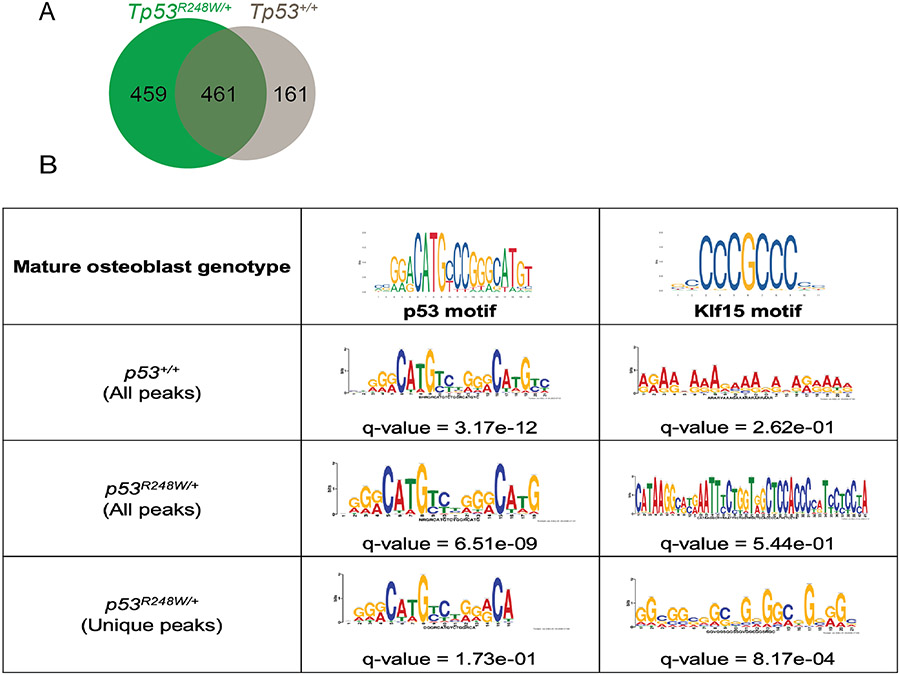
To establish significance in a human model, we engineered H1 hESCs to possess one copy of Tp53R248W, and one copy of WT Tp53. The heterozygous mutant Tp53R248W and a homozygous WT were differentiated in vitro to establish cells of an osteoblastic lineage. ChIP-seq performed for p53 identified chromatin occupancy at 774 loci in WT osteoblasts and 1147 loci in p53R248W osteoblasts (Supplementary tables 4 and 5). The increase in the number of loci with p53 chromatin occupancy support a gain of function by this missense mutant in human osteoblasts. By comparing the locations of chromatin occupancy peaks, we found 542 of the 1147 identified in p53R248W cells overlapped by at least 1bp with peaks called in WT cells, an observation that can be attributed to this heterozygous model possessing a WT Tp53 allele. The remaining 605 loci with p53 occupancy in p53R248W were unique to mutant cells.
When annotating Ensembl genes found adjacent to Tp53 chromatin occupancy peaks, WT p53 cells showed occupancy upstream of 622 genes, p53R248W cells showed occupancy upstream of 920 genes, and 461 genes had occupancy in cells of both genotypes (Figure 6A). Nearly half of the genes with p53 chromatin occupancy in p53R248W cells (459/920) show new chromatin binding not seen in WT controls. To determine whether this increased chromatin occupancy is at KLF15 binding sites, we analyzed sequences at the loci where peaks were called for motif enrichment (Figure 6B). When using sequences from all peaks, ChIP-seq of both the WT control and the heterozygous p53R248W cells demonstrate the expected finding of enriching for the p53-motif. However, only p53R248W cells enriched for the KLF15 motif and this enrichment is significant only when analysis is limited to loci uniquely occupied by mutant p53 (“Unique peaks”, Figure 6B). This experiment demonstrates that the p53R248W mutant has gained chromatin binding at the transcription factor binding sites of KLF15 in human cells of osteoblastic lineage. To establish relevance to human osteosarcoma cells, we expressed p53R175H, p53R248Q, and p53R248W mutants in the Tp53-null Saos-2 human osteosarcoma cell line and confirmed expression at the protein level (Supplementary figures 6D and 6E). Co-immunoprecipitation assays using these cells confirmed that endogenous KLF15 bound the contact mutants p53R248W and p53R248Q, but not the structural mutant p53R175H (Supplementary figure 6F). These results support mutant-specific gain of function in the form of KLF15 interaction with p53R248W/Q in human osteosarcoma, an interaction that explains increased p53R248W occupancy at KLF15 regulated genes and the activation of these genes in the tumor transcriptome.
Figure 6:
ChIP seq for Tp53 shows mutant p53R248W occupancy at peaks enriched for KLF15 motifs. A: Genes associated to ChIP peaks for mature human osteoblast cells expressing p53R248W (green) or wild-type p53. B: Enriched motifs and associated q-values within the ChIP peaks for osteoblasts.
Discussion
The p53R175H and p53R248W hotspot mutants serve as prime examples of how different mutations differentially affect the conformation of the p53 protein. Both mutations disrupt the ability of the protein to bind DNA thereby resulting in loss-of-function. However, both these proteins have gain-of-function properties that are executed through their interaction with other transcription factors, and result in tumorigenesis, increased metastasis, or resistance to drugs [1, 2]. The potencies of these two p53 missense mutants are different in different tumor types, as would be expected from structurally and functionally different proteins [22, 37]. However, mechanisms by which these p53 missense mutants execute their gain of function remain understudied. Studying these mechanisms is essential as these could lead to identification of novel vulnerabilities in mutant p53 driven cancers. We present a novel study comparing the p53R172H and the less studied p53R245W mutant in murine somatic osteosarcomas. We show that not only do the mutants drive a distinct gene expression profile from p53-null tumors, the two p53 mutants drive distinct transcriptomes leading to similar metastatic phenotypes. We also identify a repertoire of transcription factors that have enriched motifs upstream of the upregulated genes of the respective transcriptomes. The most significant transcription factors did not overlap between the two p53 mutants, suggesting that these mutants interacted with distinct sets of transcription factors to regulate unique transcriptomes that resulted in the metastatic phenotype. Although previous studies in different cancers indicate the contact mutants of p53 (p53R248W/p53R273H) to be more potent than the structural mutant (p53R175H) [22, 37], in our murine somatic osteosarcoma model, the potency was relatively similar (survival and metastatic phenotypes). However, we show that the mechanisms by which these p53 mutants result in the metastatic phenotype are completely unique.
We determined the presence of multiple transcription factor binding sites in the 10kb regions upstream of the TSSs of upregulated genes. Further, multiple transcription factors could potentially regulate the same genes, which suggested that mutant p53 may potentially bind to multiple transcription factors in the same tumor cell to regulate the differential expression of genes that mediate disease progression. We also identified novel transcription factors like EGR2, NR1D2, and E2F6 that can interact with p53R172H, and MAZ, FLI1, and SP5, that can interact with p53R245W. We identified ETS2 as a significant transcription factor interacting with p53R172H but not p53R245W. Previous studies indicate that genetic abrogation of Ets2 completely reverses the metastatic phenotype driven by p53R172H [21]. This suggests that limiting one transcription factor may be sufficient to completely abrogate a mutant p53-driven phenotype. A recent study by Xiong et al. [25], also utilized a similar approach to identify transcription factors that may potentially interact with the structural mutant p53R172H and contact mutants p53R245W and p53R270H in osteosarcomas with germline p53 mutations. The study deployed the use of algorithms that were different from those that have been presented here. Additionally, the biology of mutant p53-driven osteosarcomas may be significantly different in a germline versus a somatic setting. This may be reflected in the differences observed in the repertoire of transcription factors that interact with mutant p53 in the two studies.
KLF15 was identified as the most significant candidate that interacted with p53R245W, but not with p53R172H. KLF15 belongs to a family of 18 members, all of whom play a critical role in regulation of physiological systems. It is an important part of the transcriptional machinery in various organs like kidney, liver, heart, adipose tissue, as well as skeletal muscle [38-42]. KLF15 plays an essential role in maintaining physiological homeostasis in different organs by activation as well as repression of its target genes. However, its role in cancers remains understudied. This transcription factor seems to play contrasting roles in different cancers, suggesting that its effects in different cancer cell types may be context-specific. KLF15 suppresses breast cancer proliferation partially through upregulation of p21 and subsequent cell cycle arrest through regulation of E2F1 expression [43]. KLF15 also limits cell proliferation in gastric cancer through upregulation of p21 and p57 gene expressions and cell cycle arrest [44]. However, in lung adenocarcinoma, the high expression of KLF15 protects against apoptosis, and knockdown of Klf15 results in slower tumor growth rates, thereby suggesting an oncogenic role for KLF15 in lung adenocarcinomas [45]. Although the biology of KLF15 in skeletal tissues has been elucidated [42], effects of KLF15 on the expression of its target genes has not been explored in cancers like osteosarcomas, nor has it been studied in the context of mutant p53 gain of function. Our study indicates that KLF15 partakes in mutant p53R245W GOF, and promotes metastasis. Upon abrogation of KLF15, the metastatic potential of murine osteosarcoma cell lines expressing p53R245W decreases. Further studies are required to establish its role in somatic osteosarcoma as p53 loss of function is a common mechanism.
We identified 139 osteosarcoma patients in an MSK-IMPACT Sarcoma study [46], of which 17 had homozygous deletion of Tp53, and 23 had missense mutations in the Tp53 gene. A comparison of survival curves did not yield significant differences due to small sample sizes; however, the median survival for patients with a homozygous deletion of Tp53 was 22 months, while that of patients with a Tp53 missense mutation was 14 months, suggesting a need for considering differential therapeutic approaches based on Tp53 status. Our studies have led to the identification of KLF15 as a novel interacting partner of p53R245W, but not p53R172H in murine somatic osteosarcoma, and have highlighted the importance of considering the unique mutants of Tp53 individually for better therapeutic outcomes.
Supplementary Material
Supplementary Table 1: List of primers used in genotyping and qRT-PCR experiments.
Supplementary Table 2: List of mice belonging to different cohorts.
Supplementary Table 3: Pulmonary metastasis in p53 mutant mice.
Supplementary Table 4: Gene annotation performed by HOMER of p53 ChIP peaks from WT p53 osteoblasts.
Supplementary Table 5: Gene annotation performed by HOMER of p53 ChIP peaks from p53R248W osteoblasts.
Supplementary Table 6: Gene annotation performed by HOMER of p53 ChIP peaks from p53R248W osteoblasts.
Acknowledgements
We are grateful to the Genetically Engineered Mouse Facility, Small Animal Imaging Facility, and the Advanced Technology Genomics Core at MD Anderson Cancer Center, the Functional Genomics Core, and the Epigenomics Profiling Core at MD Anderson Cancer Center (supported by the NIH/NCI through grant P30CA016672). We would also like to thank Ms. Nikita Williams for help and guidance with tail-vein injections. CRV was supported on a fellowship from the NLM Training Program in Biomedical Informatics and Data Science (T15LM007093). AX is a CPRIT Postdoctoral Fellow in the Biomedical Informatics, Genomics, and Translational Cancer Research Training Program (BIG-TCR, CPRIT RP210045). D-FL is supported by CPRIT RR160019 and NIH/NCI R01CA246130. D-FL is a CPRIT Scholar in Cancer Research. This work was supported by Cancer Prevention and Research Institute of Texas grant RP170231 and national cancer Institute grant CA082577 to G. Lozano.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Kim MP and Lozano G, Mutant p53 partners in crime. Cell Death & Differentiation, 2018. 25(1): p. 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oren M and Rotter V, Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol, 2010. 2(2): p. a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olivier M, Hollstein M, and Hainaut P, TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol, 2010. 2(1): p. a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho Y, Gorina S, Jeffrey PD, and Pavletich NP, Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science, 1994. 265(5170): p. 346. [DOI] [PubMed] [Google Scholar]
- 5.Joerger AC, Allen MD, and Fersht AR, Crystal structure of a superstable mutant of human p53 core domain. Insights into the mechanism of rescuing oncogenic mutations. J Biol Chem, 2004. 279(2): p. 1291–6. [DOI] [PubMed] [Google Scholar]
- 6.Johnson TM, Hammond EM, Giaccia A, and Attardi LD, The p53QS transactivation-deficient mutant shows stress-specific apoptotic activity and induces embryonic lethality. Nature Genetics, 2005. 37(2): p. 145–152. [DOI] [PubMed] [Google Scholar]
- 7.Bossi G, Lapi E, Strano S, Rinaldo C, Blandino G, and Sacchi A, Mutant p53 gain of function: reduction of tumor malignancy of human cancer cell lines through abrogation of mutant p53 expression. Oncogene, 2006. 25(2): p. 304–9. [DOI] [PubMed] [Google Scholar]
- 8.Li R, Sutphin PD, Schwartz D, Matas D, Almog N, Wolkowicz R, et al. , Mutant p53 protein expression interferes with p53-independent apoptotic pathways. Oncogene, 1998. 16(25): p. 3269–77. [DOI] [PubMed] [Google Scholar]
- 9.Soussi T and Béroud C, Assessing TP53 status in human tumours to evaluate clinical outcome. Nat Rev Cancer, 2001. 1(3): p. 233–40. [DOI] [PubMed] [Google Scholar]
- 10.Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, et al. , A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell, 2009. 137(1): p. 87–98. [DOI] [PubMed] [Google Scholar]
- 11.Stambolsky P, Tabach Y, Fontemaggi G, Weisz L, Maor-Aloni R, Sigfried Z, et al. , Modulation of the vitamin D3 response by cancer-associated mutant p53. Cancer cell, 2010. 17(3): p. 273–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freed-Pastor William A., Mizuno H, Zhao X, Langerød A, Moon S-H, Rodriguez-Barrueco R, et al. , Mutant p53 Disrupts Mammary Tissue Architecture via the Mevalonate Pathway. Cell, 2012. 148(1): p. 244–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chicas A, Molina P, and Bargonetti J, Mutant p53 Forms a Complex with Sp1 on HIV-LTR DNA. Biochemical and Biophysical Research Communications, 2000. 279(2): p. 383–390. [DOI] [PubMed] [Google Scholar]
- 14.Lang GA, Iwakuma T, Suh Y-A, Liu G, Rao VA, Parant JM, et al. , Gain of Function of a p53 Hot Spot Mutation in a Mouse Model of Li-Fraumeni Syndrome. Cell, 2004. 119(6): p. 861–872. [DOI] [PubMed] [Google Scholar]
- 15.Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, et al. , Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell, 2004. 119(6): p. 847–60. [DOI] [PubMed] [Google Scholar]
- 16.Xiong S, Tu H, Kollareddy M, Pant V, Li Q, Zhang Y, et al. , Pla2g16 phospholipase mediates gain-of-function activities of mutant p53. Proceedings of the National Academy of Sciences, 2014. 111(30): p. 11145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noone A-M, Cronin KA, Altekruse SF, Howlader N, Lewis DR, Petkov VI, et al. , Cancer Incidence and Survival Trends by Subtype Using Data from the Surveillance Epidemiology and End Results Program, 1992–2013. Cancer Epidemiology Biomarkers & Prevention, 2017. 26(4): p. 632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Broadhead ML, Clark JCM, Myers DE, Dass CR, and Choong PFM, The Molecular Pathogenesis of Osteosarcoma: A Review. Sarcoma, 2011. 2011: p. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Overholtzer M, Rao PH, Favis R, Lu XY, Elowitz MB, Barany F, et al. , The presence of p53 mutations in human osteosarcomas correlates with high levels of genomic instability. Proc Natl Acad Sci U S A, 2003. 100(20): p. 11547–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walkley CR, Qudsi R, Sankaran VG, Perry JA, Gostissa M, Roth SI, et al. , Conditional mouse osteosarcoma, dependent on p53 loss and potentiated by loss of Rb, mimics the human disease. Genes & development, 2008. 22(12): p. 1662–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pourebrahim R, Zhang Y, Liu B, Gao R, Xiong S, Lin PP, et al. , Integrative genome analysis of somatic p53 mutant osteosarcomas identifies Ets2-dependent regulation of small nucleolar RNAs by mutant p53 protein. Genes & Development, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Xiong S, Liu B, Pant V, Celii F, Chau G, et al. , Somatic Trp53 mutations differentially drive breast cancer and evolution of metastases. Nature Communications, 2018. 9(1): p. 3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, and Berns A, Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nature Genetics, 2001. 29(4): p. 418–425. [DOI] [PubMed] [Google Scholar]
- 24.Bailey TL, Johnson J, Grant CE, and Noble WS, The MEME Suite. Nucleic Acids Research, 2015. 43(W1): p. W39–W49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiong S, Chachad D, Zhang Y, Gencel-Augusto J, Sirito M, Pant V, et al. , Differential Gain-of-Function Activity of Three p53 Hotspot Mutants In Vivo. Cancer Res, 2022. 82(10): p. 1926–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Do PM, Varanasi L, Fan S, Li C, Kubacka I, Newman V, et al. , Mutant p53 cooperates with ETS2 to promote etoposide resistance. Genes Dev, 2012. 26(8): p. 830–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim MP, Li X, Deng J, Zhang Y, Dai B, Allton KL, et al. , Oncogenic KRAS Recruits an Expansive Transcriptional Network through Mutant p53 to Drive Pancreatic Cancer Metastasis. Cancer Discovery, 2021. 11(8): p. 2094–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gencel-Augusto J, Su X, Qi Y, Whitley EM, Pant V, Xiong S, et al. , Dimeric p53 Mutant Elicits Unique Tumor-Suppressive Activities through an Altered Metabolic Program. Cancer Discovery, 2023: p. OF1–OF20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu A, Zhou R, Tu J, Huo Z, Zhu D, Wang D, et al. , Establishment of a human embryonic stem cell line with homozygous TP53 R248W mutant by TALEN mediated gene editing. Stem Cell Res, 2018. 29: p. 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tu J, Huo Z, Yu Y, Zhu D, Xu A, Huang M-F, et al. , Hereditary retinoblastoma iPSC model reveals aberrant spliceosome function driving bone malignancies. Proceedings of the National Academy of Sciences, 2022. 119(16): p. e2117857119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee DF, Su J, Kim HS, Chang B, Papatsenko D, Zhao R, et al. , Modeling familial cancer with induced pluripotent stem cells. Cell, 2015. 161(2): p. 240–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou R, Xu A, Tu J, Liu M, Gingold JA, Zhao R, et al. , Modeling Osteosarcoma Using Li-Fraumeni Syndrome Patient-derived Induced Pluripotent Stem Cells. J Vis Exp, 2018(136). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaseniit KE, Katz N, Kolber NS, Call CC, Wengier DL, Cody WB, et al. , Modular, programmable RNA sensing using ADAR editing in living cells. Nat Biotechnol, 2022. [DOI] [PubMed] [Google Scholar]
- 34.Kager L, Zoubek A, Pötschger U, Kastner U, Flege S, Kempf-Bielack B, et al. , Primary metastatic osteosarcoma: presentation and outcome of patients treated on neoadjuvant Cooperative Osteosarcoma Study Group protocols. Journal of clinical oncology, 2003. 21(10): p. 2011–2018. [DOI] [PubMed] [Google Scholar]
- 35.Post SM, Quintás-Cardama A, Terzian T, Smith C, Eischen CM, and Lozano G, p53-dependent senescence delays Emu-myc-induced B-cell lymphomagenesis. Oncogene, 2010. 29(9): p. 1260–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Terzian T, Suh YA, Iwakuma T, Post SM, Neumann M, Lang GA, et al. , The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss. Genes Dev, 2008. 22(10): p. 1337–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hassin O, Nataraj NB, Shreberk-Shaked M, Aylon Y, Yaeger R, Fontemaggi G, et al. , Different hotspot p53 mutants exert distinct phenotypes and predict outcome of colorectal cancer patients. Nature Communications, 2022. 13(1): p. 2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anzai K, Tsuruya K, Ida K, Kagawa T, Inagaki Y, and Kamiya A, Kruppel-like factor 15 induces the development of mature hepatocyte-like cells from hepatoblasts. Scientific Reports, 2021. 11(1): p. 18551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gray S, Feinberg MW, Hull S, Kuo CT, Watanabe M, Banerjee SS, et al. , The Krüppel-like Factor KLF15 Regulates the Insulin-sensitive Glucose Transporter GLUT4*. Journal of Biological Chemistry, 2002. 277(37): p. 34322–34328. [DOI] [PubMed] [Google Scholar]
- 40.Rane MJ, Zhao Y, and Cai L, Krϋppel-like factors (KLFs) in renal physiology and disease. EBioMedicine, 2019. 40: p. 743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang B, Haldar SM, Lu Y, Ibrahim OA, Fisch S, Gray S, et al. , The Kruppel-like factor KLF15 inhibits connective tissue growth factor (CTGF) expression in cardiac fibroblasts. Journal of Molecular and Cellular Cardiology, 2008. 45(2): p. 193–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zakeri S, Aminian H, Sadeghi S, Esmaeilzadeh-Gharehdaghi E, and Razmara E, Krüppel-like factors in bone biology. Cellular Signalling, 2022. 93: p. 110308. [DOI] [PubMed] [Google Scholar]
- 43.Yoda T, McNamara KM, Miki Y, Onodera Y, Takagi K, Nakamura Y, et al. , KLF15 in breast cancer: a novel tumor suppressor? Cellular Oncology, 2015. 38(3): p. 227–235. [DOI] [PubMed] [Google Scholar]
- 44.Sun C, Ma P, Wang Y, Liu W, Chen Q, Pan Y, et al. , KLF15 Inhibits Cell Proliferation in Gastric Cancer Cells via Up-Regulating CDKN1A/p21 and CDKN1C/p57 Expression. Digestive Diseases and Sciences, 2017. 62(6): p. 1518–1526. [DOI] [PubMed] [Google Scholar]
- 45.Gao L, Qiu H, Liu J, Ma Y, Feng J, Qian L, et al. , KLF15 promotes the proliferation and metastasis of lung adenocarcinoma cells and has potential as a cancer prognostic marker. Oncotarget, 2017. 8(66): p. 109952–109961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nacev BA, Sanchez-Vega F, Smith SA, Antonescu CR, Rosenbaum E, Shi H, et al. , Clinical sequencing of soft tissue and bone sarcomas delineates diverse genomic landscapes and potential therapeutic targets. Nat Commun, 2022. 13(1): p. 3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: List of primers used in genotyping and qRT-PCR experiments.
Supplementary Table 2: List of mice belonging to different cohorts.
Supplementary Table 3: Pulmonary metastasis in p53 mutant mice.
Supplementary Table 4: Gene annotation performed by HOMER of p53 ChIP peaks from WT p53 osteoblasts.
Supplementary Table 5: Gene annotation performed by HOMER of p53 ChIP peaks from p53R248W osteoblasts.
Supplementary Table 6: Gene annotation performed by HOMER of p53 ChIP peaks from p53R248W osteoblasts.
Data Availability Statement
The data generated in this study are publicly available in National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) at GSE231852. All other raw data are available upon request from the corresponding author.