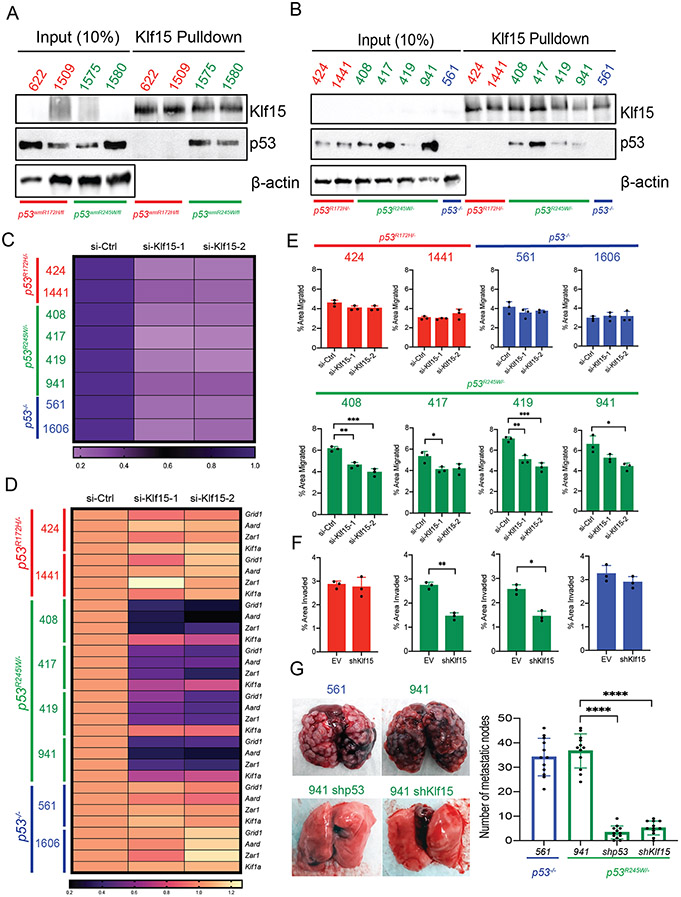
Figure 5:
KLF15 selectively binds to p53R245W but not p53R172H to execute mutant p53 gain of function. A: Co-immunoprecipitation assays in primary murine osteosarcomas expressing stable p53R172H (622, 1509 – in red) or p53R245W (1575, 1580 – in green) mutants. Pulldown with KLF15 antibody (top) and probe with p53 antibody (middle); β-actin served as loading control (bottom). B: Co-immunoprecipitation assay in murine osteosarcoma cell lines with stable p53R172H (424, 1441 – in red) or p53R245W (408, 417, 419, 941 – in green) mutants or p53-null cell line (561 – in blue). Pulldown with KLF15 antibody (top) and probed with p53 antibody (middle); β-actin served as loading control (bottom). C: Expression of Klf15 upon treatment with two independent siRNAs against Klf15 as quantified by qRT-PCR in murine OS cell lines expressing p53R172H (in red), p53R245W (in green) mutants or no p53 (561, 1606 – in blue); Gapdh used as control. D: Gene expression analysis by qRT-PCR in murine osteosarcoma cell lines for transcriptional targets of KLF15 and p53R245W – Grid1, Aard, Zar1. Kif1a is a gene target of p53R245W but is not a transcriptional target of KLF15 and was used as a negative control; Gapdh served as control. E: Effects of Klf15 knockdown on migration potential of murine osteosarcoma cell lines. F: Effects of Klf15 knockdown on invasion potential on murine osteosarcoma cell lines expressing p53R172H (red), p53R245W (green) or null for p53 (blue). G: Mouse lungs with metastatic nodes after tail-vein injections of p53-null cells (blue) or cells expressing p53R245W (green), with a stable knockdown of p53R245W, or stable knockdown of Klf15 (green) quantified on the right. Significance calculated by student’s t-test from experiments using at least 3 biological replicates; *p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.001.