Figure 5.
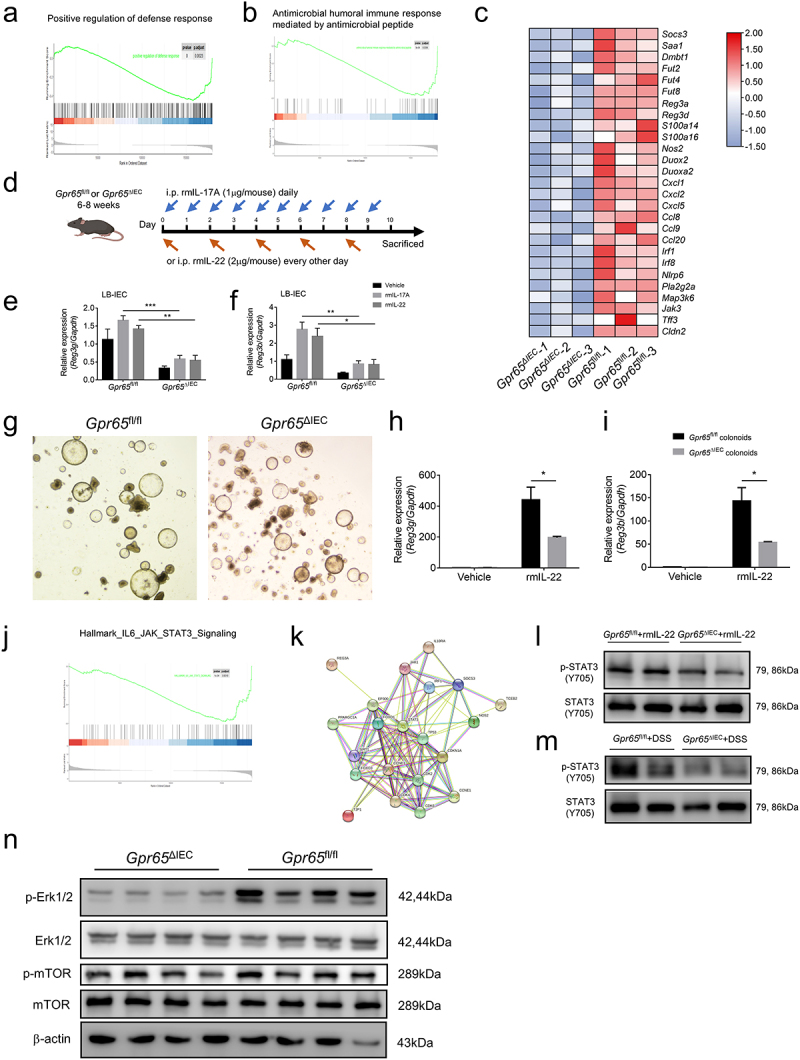
GPR65 deficiency drives distinct IEC transcriptional programs and compromises the downstream STAT3 signaling. colonic IECs were isolated from Gpr65ΔIEC mice and Gpr65fl/fl littermates for RNA sequencing analysis. (a, b) GSEA analysis. (c) Heatmap of DEGs related to IEC antimicrobial gene profiles. (d) a schematic overview of the AMP induction assay by systemic injection with rmIL-17A or rmIL-22. (e, f) qPCR analysis of relative mRNA expression of Reg3g and Reg3b. (g) representative microscopical photographs of colonoids from Gpr65ΔIEC and littermate Gpr65fl/fl mice on day 10 of culture. Original magnification: × 40. (h, i) the colonoids from Gpr65ΔIEC and Gpr65fl/fl mice were stimulated with or without rmIL-22 (100 ng/mL) for 24 h. The relative mRNA expression of Reg3g and Reg3b was detected by qPCR analysis. (j) GSEA analysis for Hallmark_JAK_STAT3_Signaling. (k) STAT3-focused interaction network of genes downregulated by GPR65 signaling. (l) colonic IECs were isolated from Gpr65ΔIEC mice and Gpr65fl/fl littermates, and stimulated with rmIL-22 (100 ng/mL) for 15 min. The protein levels of p-STAT3 and STAT3 were determined by immunoblotting analysis. (m) immunoblotting analysis of p-STAT3 and STAT3 in the colonic IECs from the indicated groups of mice. (n) immunoblotting analysis of p-Erk1/2, Erk1/2, p-mTOR, mTOR, and β-actin in the colonic IECs from the indicated groups of mice. *p < 0.05, **p < 0.01, ***p < 0.001. Data are representative of three independent experiments.
