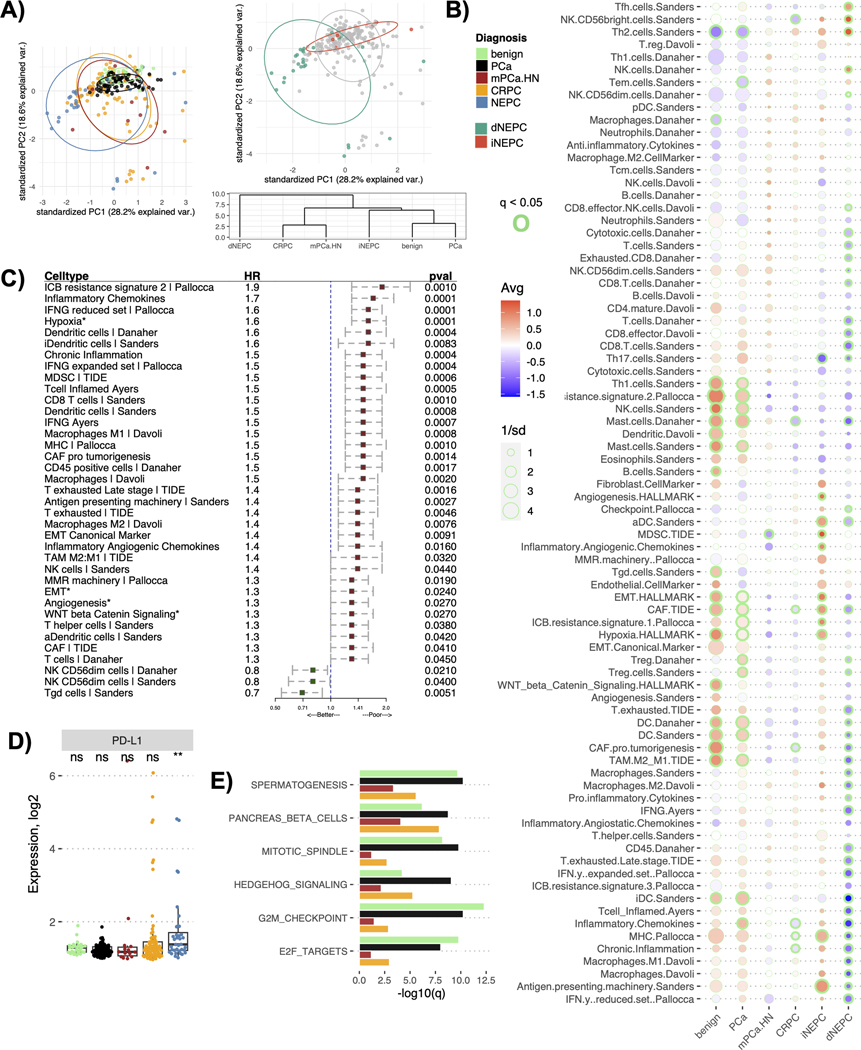
Figure 3.
TME profile of distinct prostate cancer types in the cohort. A) Plot for the principal component analysis applied to the TME profiles (comprised of 80 gene signatures) of the prostate cancer cohort colored either by diagnosis (left panel) or by NEPC immune classes (right panel top). The right panel bottom shows a cluster dendrogram for the prostate cancer diagnosis based on their averaged TME profiles. The dendrogram was constructed using the hierarchical clustering (distance= euclidean, method=ward D2), B) Bubble plot showing differences in the averaged TME profiles among the prostate cancer diagnosis in the cohort. The color of the circles indicates the mean (Avg) enrichment score by diagnosis for that feature, with red denoting high values and blue denoting low values on a color gradient scale. The size of the circle is determined by 1/standard deviation (sd) of the scores within that diagnosis, larger the circle smaller the sd among the tumors for that feature scores. The enrichment scores significantly different from at least four other cancers diagnoses being compared at a q < 0.05 are outlined with green, the thickness of the circle denotes the number of diagnoses it is significantly different from, C) Forest plots showing associations between the OS and individual features of the TME profile. Asterisk indicates that the corresponding feature is a cancer hallmark pathway. Hazard ratio (HR) and p-values (pval) are reported from the Cox proportional-hazards regression models adjusted for cancer type. The plot only shows associations that were significant at p < 0.05, D) Boxplot showing the differences in the gene expression levels of PD-L1 among the prostate cancer types. Asterisk denotes levels significantly different from all other cancer types compared (p < 0.05), while ns denotes not significant p-values from Wilcoxon signed-rank test, E) Barplot showing the cancer hallmark pathways significantly overexpressed in NEPCs compared to all other prostate diagnosis. The x-axis shows q values (-log10 scale) for the differences between the enrichment scores for the corresponding pathways in NEPC versus the cancer type being compared to.