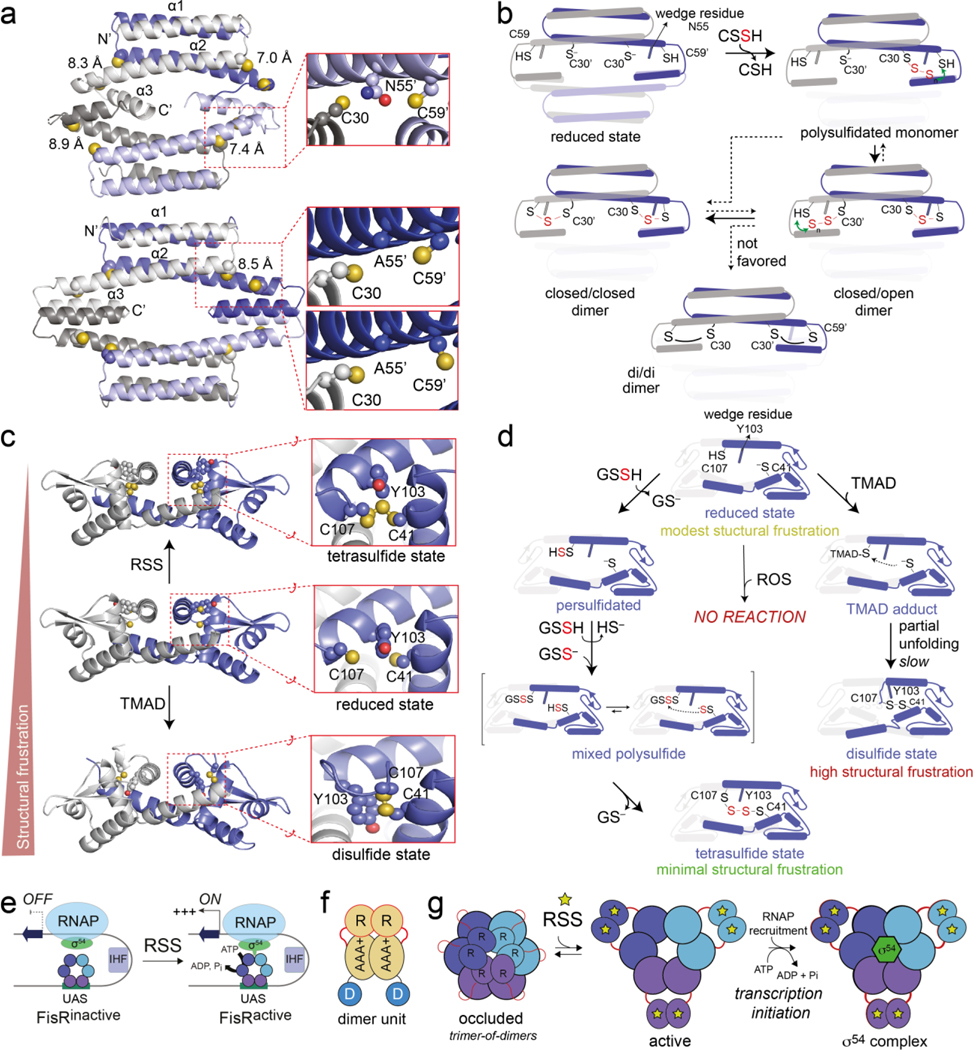
Figure 4.
(a) X-ray crystal structures of C9A (upper) and C9A/N55A S. pneumoniae CstRs, with dithiol sensing sites highlighted in the inset panels [69]. (b) A summary of the results of kinetically resolved native mass spectrometry-based profiling of reaction products when reduced CstR is incubated with a molar excess of CSSH. Only one of the dimer units of the CstR tetramer are shown in the foreground for clarity. These five states shown are not representative of discrete intermediates, but instead capture collections of structures that conform to the indicated trivial designation, e.g., “closed/closed” represents species that harbor tri- or tetrasulfide linkages on both sides of the dimer, not just the doubly trisulfidated species as shown [69,70]. (c) X-ray crystal structures of three distinct states of the RcSqrR dimer, with one of the two dithiol sensing sites highlighted in the expanded view [17]. (d) A cartoon summary of kinetically resolved native mass spectrometry-based reaction products when reduced SqrR is reacted with a molar excess of GSSH [17]. Reactivity of only one of the protomers of the SqrR dimer are shown in the foreground for clarity. (e) Generic cartoon of the regulatory mechanism of hexameric AAA+ σ54-dependent transcriptional activators like FisR. UAS, upstream activation sequence; IHF, integration host factor; RNAP, RNA polymerase. (f) The fundamental functional unit of FisR is a dimer, where R, AAA+ and D correspond to the N-terminal regulatory domain, the catalytic ATPase domain and the DNA binding domain, which engages the UAS, respectively. (g) One of a number of possible regulatory models for a FisR, with the nature of the RSS-sensing mechanism not broadly established, but in one case appears to involve transpersulfidation of the R domain directly [16].