Figure 1. Detection of SARS-CoV-2 in placentae by histology and bulk RT-qPCR.
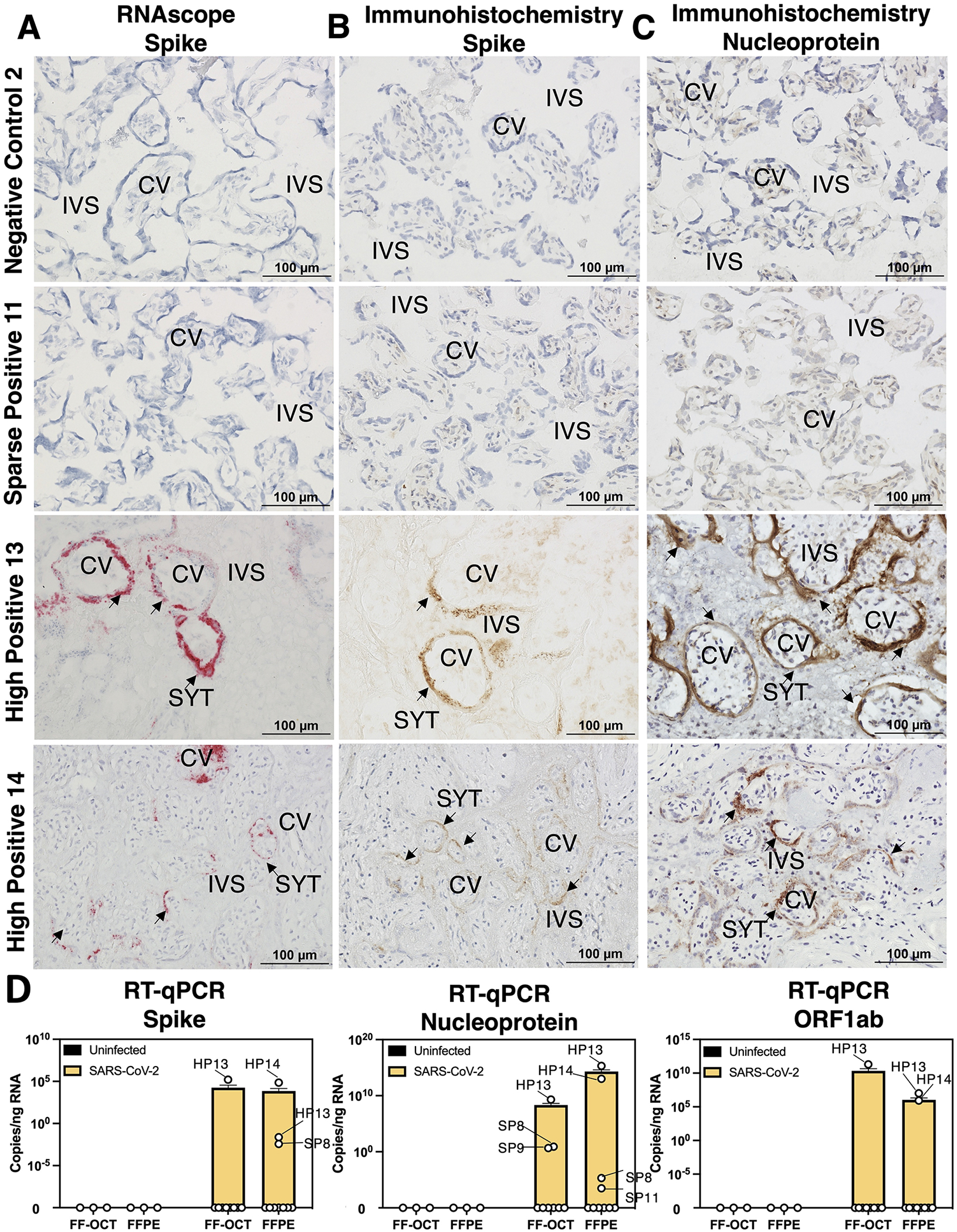
(A-C) Fresh-frozen in optimal cutting temperature serum (FF-OCT) tissue blocks from spatial transcriptomics samples were cryosectioned and subject to (A) RNA in situ probing for SARS-CoV-2 Spike (S), (B) immunohistochemistry (IHC) for S, and (C) IHC staining for SARS-CoV-2 Nucleocapsid (N). Images were taken at 20x magnification and each row represents images obtained from an individual participant. (D) FF-OCT and formalin-fixed and paraffin-embedded (FFPE) blocks were subject to reverse transcription and quantitative polymerase chain reactions (RT-qPCR) probing for S, N, or ORF1ab SARS-CoV-2 transcripts. Based on these results, placentae were grouped for analysis into negative controls (NC), maternal positive but SARS-CoV-2 was not detected in the placenta (ND), sparse positive (SP) if SARS-CoV-2 was detected by RT-qPCR where ct values <27 were observed (limit of detection = 1/7,000 cells) or ≥1 SARS-CoV-2 transcripts per spot were observed spatial transcriptomics (limit of detection = 1/661 cells), and high positive (HP) where RT-qPCR ct values <15 and ≥2–1,554 SARS-CoV-2 transcripts per spot were observed. Abbreviations: CV= chorionic villi, IVS= intervillous space, and SYT= syncytiotrophoblast.
