Figure 7. Caution in assigning fetal sex differences in gene expression associated with placental SARS-CoV-2 without spatial resolution.
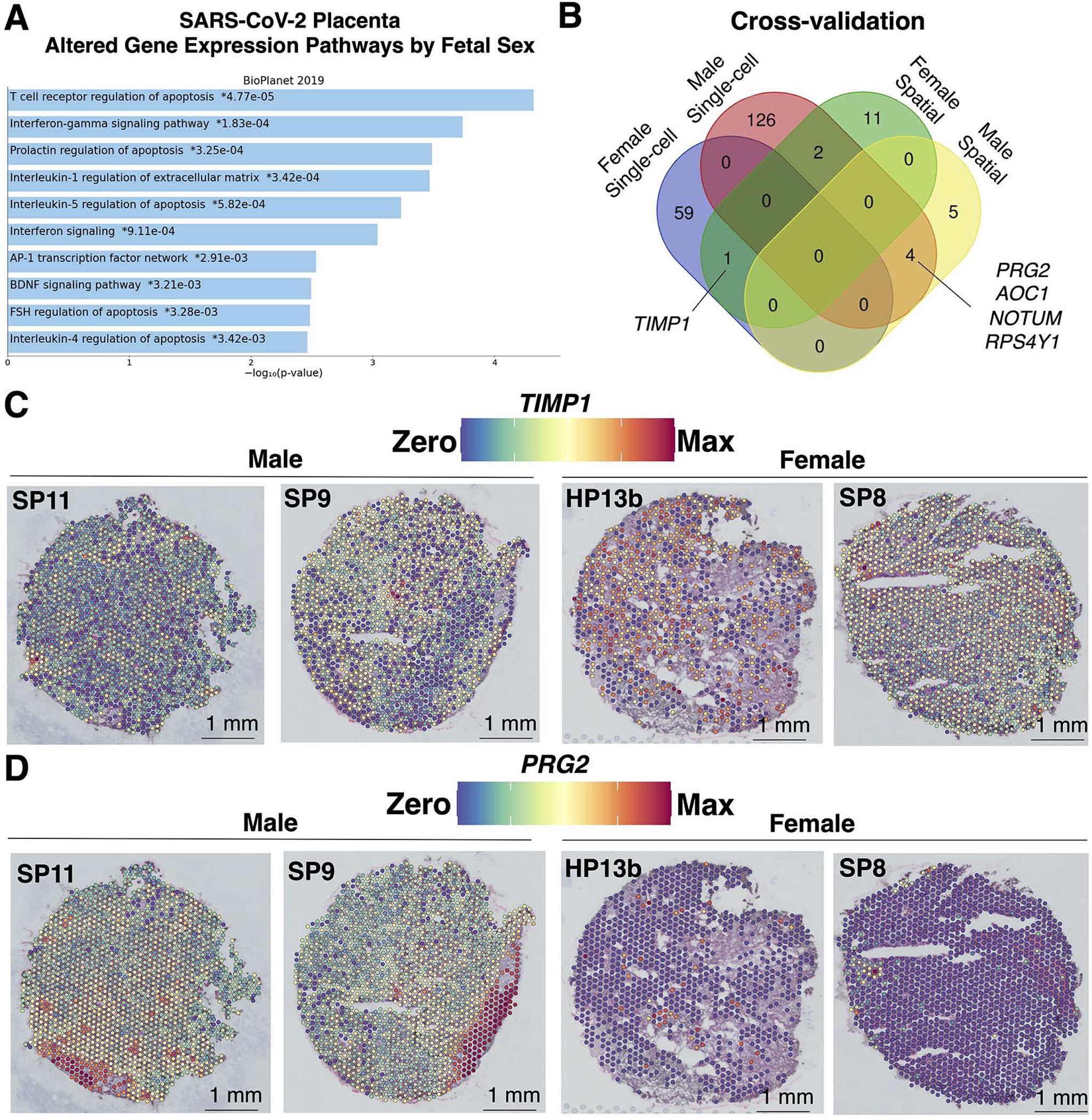
(A) The 208 significantly (q<0.05) differentially expressed genes in the SARS-CoV-2 scRNA-seq (147,906 transcriptomes; n=15 female and 30 male19 and spatial transcriptomics (9,446 transcriptomes; n=4 male and 5 female) data based on fetal sex were uploaded to EnrichR for BioPlanet pathway analysis. The top 90th-quartile of most significant (q<0.05) pathways were plotted. (B) The 208 genes with sex differences in expression from the SARS-CoV-2 scRNA-seq and spatial transcriptomics datasets were compared and plotted as a Venn diagram, revealing 4 male and 1 female cross-validated genes. (C-D) Spatial gene expression of cross-validated genes (C) TIMP1 and (D) PRG2 were upregulated in the villous space in females and maternal decidua regions of male SARS-CoV-2 placentae, respectively.
