Figure 1. Mycobacteriophages with glycosylated virions.
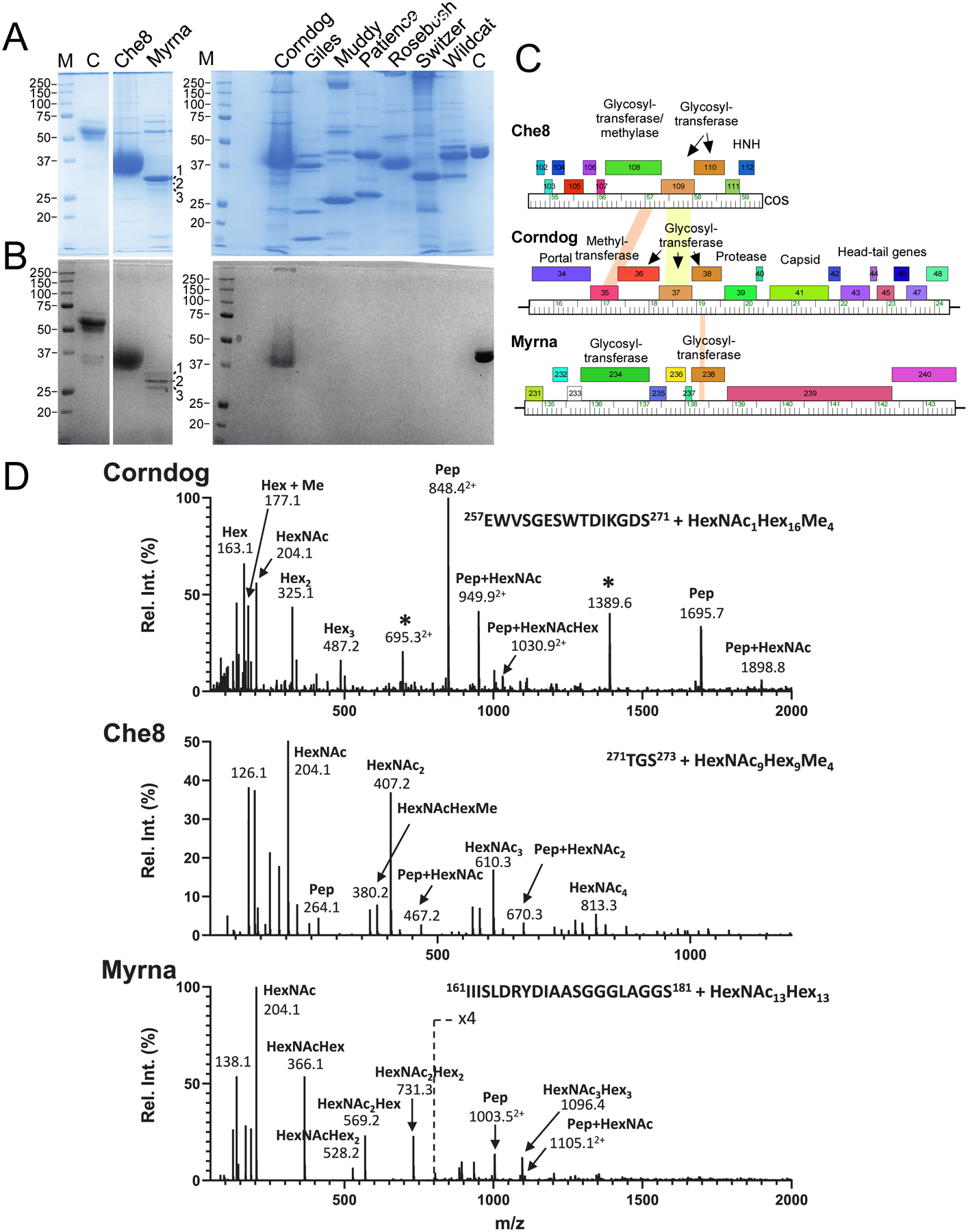
A and B. SDS-PAGE analysis of mycobacteriophage virions. Replicate gels were stained with either Coomassie Blue (A) or glycostain (B). Molecular weight markers (M), a glycostain positive control (C), and phages as labeled are shown. The major glyco-stained bands in Che8 and Corndog and the three Myrna glyco-stained bands labeled 1, 2, and 3 were excised for analysis by MS/MS. C. Genome segments of phages Che8, Corndog, and Myrna encoding glycosyltransferase genes. Genes are shown as colored boxes above the genome ruler and putative functions are indicated. D. Glycopeptide CID-MS/MS spectra from the tail tube subunit of Corndog (gp49, top), the capsid subunit from Che8 (gp6, middle) and a minor capsid subunit of Myrna (gp98, bottom) from band 1; the precursor ions were m/z 1516.63+, m/z 1202.83+ and m/z 1351.45+, respectively (underlined in Fig. S1). Glycan oxonium ions (m/z 163.1, 204.1, 325.2, 366.1, etc.) are present in the lower half of the MS/MS spectra. Partial methylation of hexoses in Corndog and Che8 is evident by the presence of the oxonium ions at m/z 177.1, 339.2 and 380.2. Fragment ions corresponding to the peptide (Pep) and the peptide linked to HexNAc (Pep+HexNAc) are identified in all the MS/MS spectra. Ions marked with an asterisk in the Corndog spectrum are derived from a co-eluting peptide and are un-related to the glycopeptide under investigation. Details of the MS analyses are in Supplementary Data Set 1. HexNAc, N-Acetylhexosamine, Hex, unmodified hexose, Me, methyl.
