Figure 2. Che8 110 is required for glycosylation.
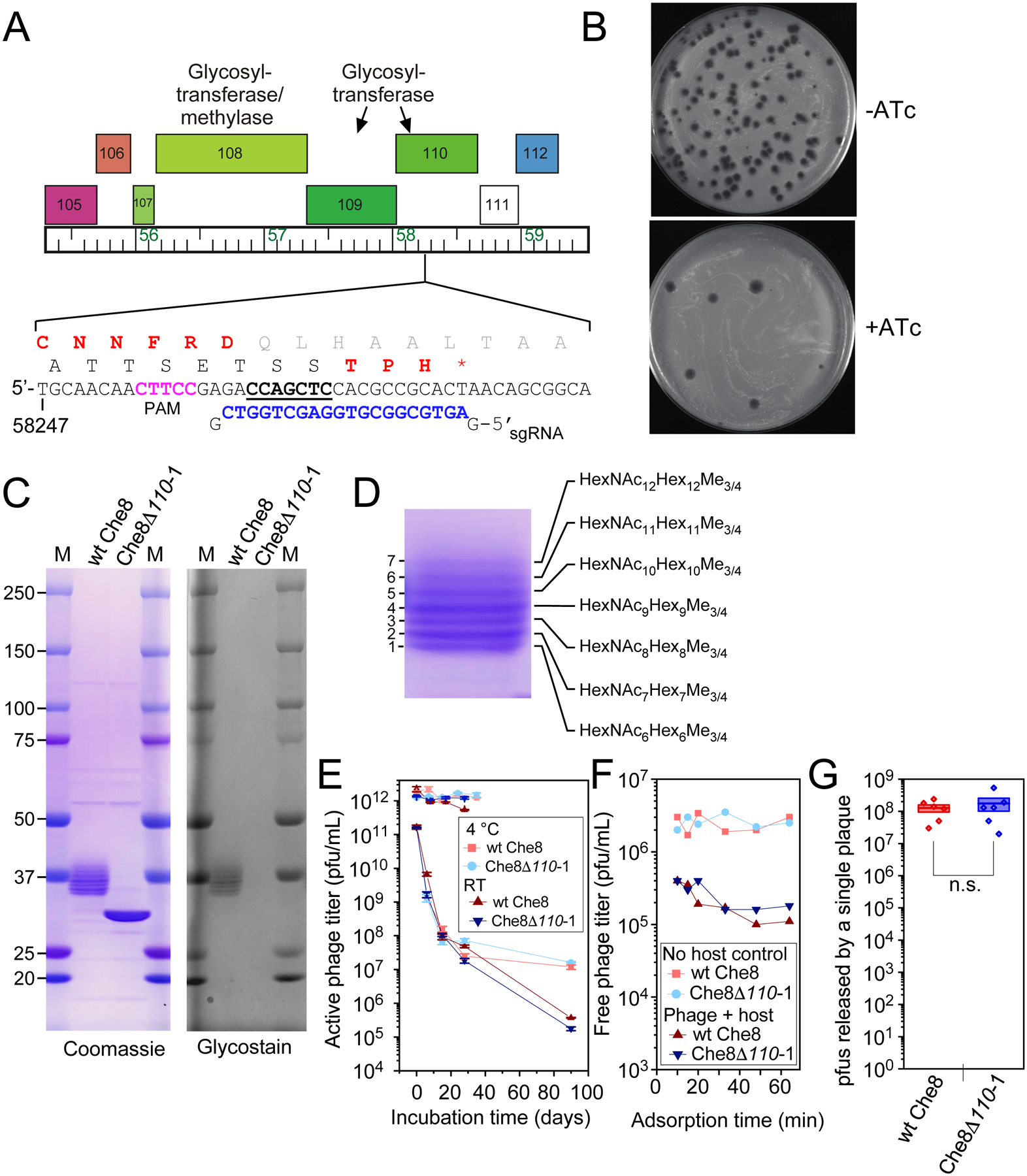
A and B. A sgRNA-expressing plasmid was designed to target Che8 gene 110 encoding a putative N-acetyl-galactosyl transferase. When sgRNA and Cas9 expression is induced (+ATc) the efficiency of plaquing of Che8 is reduced approximately two orders of magnitude (panel B). Candidate CRISPR-escape mutants were isolated and one was shown to contain a seven base pair deletion (underlined) within the sequence targeted by the sgRNA (blue type). The reading frame shift produces a truncated variant of gp110 (amino acids in red type). C. SDS-PAGE of Che8 and Che8Δ110-1 virions, stained with either Coomassie Blue or Glycostain as indicated. Seven bands are resolved in the 37 kDa size range in wild type Che8, all of which are glycosylated. In Che8Δ110-1 these collapse to a single band containing both the major capsid (29.04 kDa) and tail tube subunits (29.78 kDa), both of which are unglycosylated. D. An expanded view of the glycosylated Che8 proteins, and a schematic of the arrangements of sugars in each band. HexNAc, N-Acetylhexosamine, Hex, unmodified hexose, Me, methyl. Further mass spectrometry analysis is shown in Figure S1 and in Supplementary Data Set 2. E. Viability of wild type and mutant Che8 particles with titers starting at either 1011 or 1012 pfu/mL after prolonged storage at room temperature or 4 °C. Averages of two or three technical replicates and standard error bars are shown. F. Adsorption of wild type and mutant Che8 to M. smegmatis. G. Fecundity of wild type and mutant Che8 as determined by the number of infectious particles in individual plaques. Six technical replicates are shown as individual datapoints on top of a box plot indicating the mean (central line) +/− one standard error (box boundaries).
