Fig. 5.
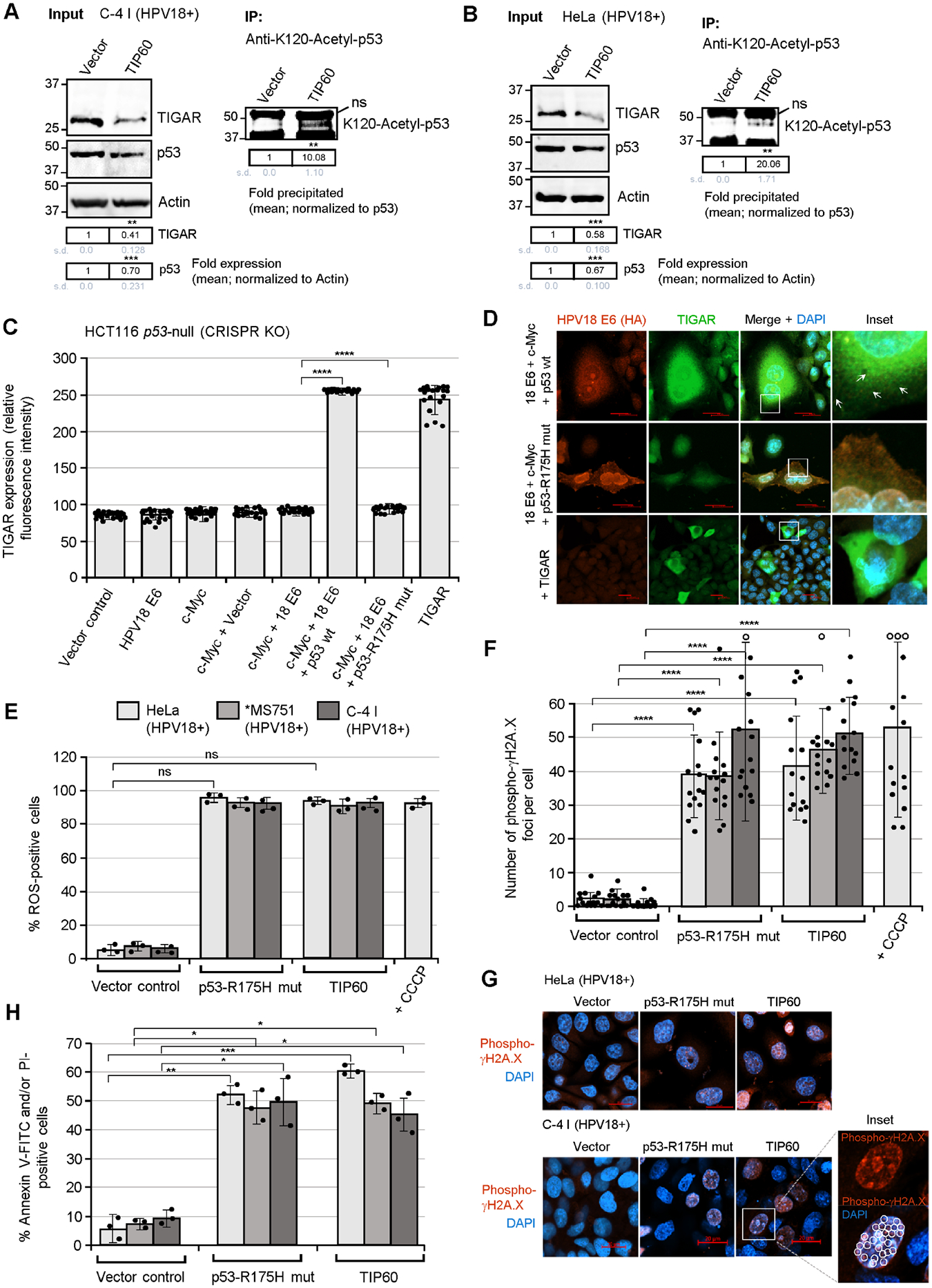
The activation of TIGAR by the E6 oncoprotein is dependent upon p53 functions and the inhibition of p53 lysine K120-acetylation in hrHPV-transformed cells. (A and B) To determine if the inhibition of TIP60-mediated lysine K120-acetylation of p53 is required for the expression of TIGAR in hrHPV-transformed cells, the HPV18+ C-4 I (in A) and HeLa (in B) cell-lines were transfected with either an expression construct for wild-type TIP60 (HA-tagged) or an empty CβS vector as negative control. The overexpression of TIP60 (HA) was confirmed by immunofluorescence-microscopy and immunoblotting and is shown in Figs. S8B and S8C. The input levels of TIGAR and p53 were quantified and normalized relative to Actin expression. The K120-acetylated p53 protein was immunoprecipitated using a monoclonal Anti-K120-Acetyl-p53 antibody (Abcam) and Protein G-agarose quantified with normalization for the relative input levels of p53 (right panels). (C and D) To determine whether p53 is required for the expression and mitochondrial targeting of TIGAR by the viral E6 oncoproteins, the p53-null human carcinoma cell-line, HCT116 p53−/− (which contains a homozygous CRISPR-knockout of the p53 gene; Horizon Discovery), was transfected with various expression constructs for c-Myc, HPV18 E6 (HA), wild-type p53, or a dominant-negative p53-R175H DNA-binding mutant, and immunofluorescence-confocal microscopy was performed to quantify and visualize the TIGAR protein in punctate mitochondrial structures (see arrows in D). The relative fluorescence intensity of the TIGAR-specific signal was measured using ZEN OS imaging software and graphed in C. The expression of wild-type p53 in the HCT116 p53-null cells cotransfected with expression constructs for HPV18 E6, c-Myc, and p53 was detected by immunoblotting and is shown in Fig. S8A. (E) The effects of the dominant-negative p53-R175H mutant and overexpression of TIP60 upon the accumulation of damaging ROS in hrHPV-transformed cells were assessed by transfecting the HPV18+ cell-lines, HeLa, MS751, and C-4 I, with CMV-p53-R175H, pOZ-TIP60 (HA), or an empty CβS vector control and then staining the cells with a fluorescent chemical ROS probe, CellROX-Deep Red, followed by confocal microscopy analysis. (F and G) The HPV18+ cell-lines, HeLa, MS751, and C-4 I, were transfected as described in E and then immunostained using an Anti-Phospho-γH2A.X primary antibody to visualize the recruitment of this phosphorylated histone variant to sites of DNA-damage. HeLa cells treated with CCCP were included as a positive control. Immunofluorescence-confocal microscopy was performed to quantify the relative numbers of phospho-JH2A.X foci (red signal in G) per cell. The open circles indicate data out-of-range. DAPI nuclear-staining (blue signal in G) is provided for reference. An enlarged inset area shows several phospho-γH2A.X-positive punctate structures (circles) within the nucleus of a C-4 I cell transfected with the pOZ-TIP60 expression construct. Scale bar, 20 μm. (H) The requirement for p53 functions and the effects of TIP60 overexpression upon cellular apoptosis in hrHPV-transformed cells were determined by transfecting HPV18+ cell-lines (HeLa, MS751, and C-4 I) with CMV-p53-R175H, pOZ-TIP60 (HA), or an empty CβS vector and then staining the cells with Annexin V-FITC and PI (BD-Pharmingen). A DIC filter was used to visualize all cells in the merged images. Data in C, E, F, and H is mean ± SD. N-value = 3. The asterisks denote statistical significance as determined using unpaired two-tailed Student’s t-tests (*P<0.0332, **P<0.0021, ***P<0.0002, ****P<0.0001, not significant (ns) 0.1234).
