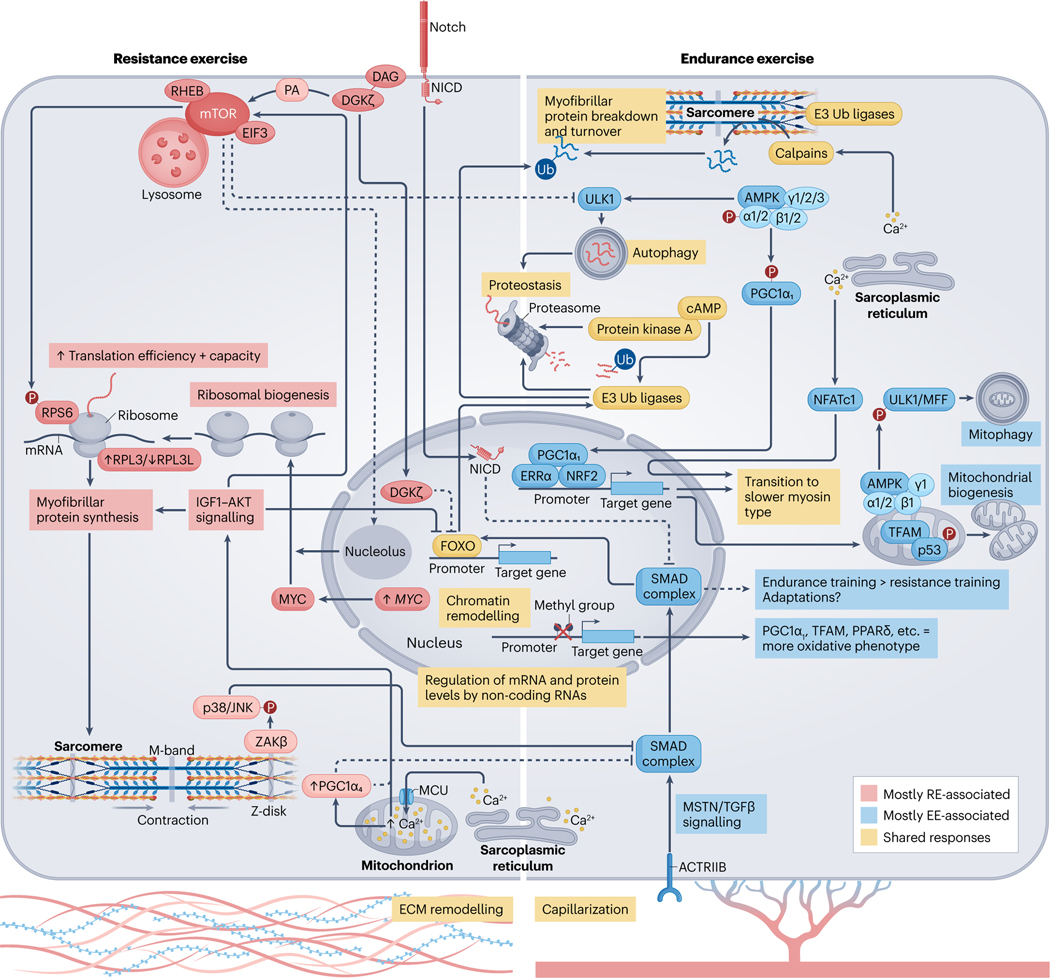
Fig. 3 |. Molecular responses to acute exercise and exercise training.
Exercise-induced alterations in circulating molecules19,124 and the intramuscular milieu55,101, together with mechanical tension178, initiates a temporal series of biochemical and molecular events that lead to muscle adaptation. Activation of signalling cascades promote substantial posttranslational modification of the muscle proteome90,170 and DNA accessibility198,199,230. Collectively, this drives transcription factor-dependent169 changes in gene expression167,183, alongside microRNAs204 and long-non-coding RNAs205 that are thought to ‘fine-tune’ the molecular responses to exercise. Endurance exercise (EE) and resistance exercise (RE) are often considered divergent stimuli, primarily driving oxidative versus hypertrophic muscle adaptations, respectively. However, common processes among exercise modalities can result in shared enrichment of signalling cascades90 and transcriptional networks183 in the post-exercise period. For example, coordinated proteolysis is detected following acute exercise, irrespective of exercise type90. This may require cAMP–protein kinase A (PKA)190,191 and ensures protein quality control and physiological muscle remodelling. 5′-AMP-activated protein kinase (AMPK) activity90 and the expression of total PGC1A mRNA183 are also increased after a single bout of either endurance or resistance exercise. AMPK phosphorylation activates peroxisome proliferator-activated receptor- coactivator isoform 1 ()279, and both AMPK and potentiate angiogenic factors260,280 and mitochondrial bioenergetics48,235,260,279 in muscle. After endurance exercise, a distinct pool of mitochondrial AMPK (composed of , , and isoforms) regulates mitophagy187–189 and promoter hypomethylation facilitates the transcription of peroxisome proliferator-activated receptor- () and mitochondrial transcription factor A (TFAM)230. Whether resistance exercise elicits these same effects is unclear. Despite similarities, there are more distinct than overlapping post-exercise responses between modalities90,183. Rapamycin-sensitive substrates of mammalian target of rapamycin (mTOR) complex 1 (mTORC1) are phosphorylated to a greater extent after resistance exercise90. Mechanical overload initiates translocation of the mTOR–lysosomal complex174 and diacylglycerol (DAG) kinase- ()173 to the sarcolemma. Here, mTOR colocalization with RAS homologue enriched in brain (RHEB) and eukaryotic initiation factor 3 (EIF3)174 and phosphatidic acid (PA) produced by 173 might coalesce to fully stimulate mTORC1 (ref. 175) and the translation of contraction-associated mRNAs. Acute attenuation of UNC-51-like autophagy activating kinase 1 (ULK1) autophagic signalling after resistance exercise65 and nuclear -mediated suppression of forkhead box protein O (FOXO)-dependent proteasomal degradation could also support muscle mass by moderating the breakdown of myofibrillar proteins173. At the sarcomere, contraction recruits to the Z-disc179 where it acts through JUN N-terminal kinase 1 (JNK1) and JNK2 (refs. 179,181), potentially alongside Notch182, to inhibit myostatin (MSTN)/transforming growth factor- () signalling. This represents one of many intracellular changes permitting resistance training-induced hypertrophy over endurance-like adaptations181. The upregulation of MYC with resistance exercise stimulates ribosomal biogenesis180,198, and the formation of a specialized pool of ribosomes with a high ratio of ribosomal protein large 3 (RPL3) to RPL3-like (RPL3L)238 may favour protein synthesis over translational fidelity. MYC expression is mTOR-independent but MYC cooperation with mTOR is necessary to successfully increase ribosomal content180, possibly requiring an mTOR-driven reorganization of nucleoli to aid rRNA transcription281. Divergence between endurance and resistance exercise at the transcriptomic level is magnified after a period of training183. Endurance training increases electron transport chain complex expression183, mitochondrial content and muscle oxidative capacity12. Conversely, growth-related pathways183, ribosomal abundance198 and muscle mass12 are augmented more by resistance training. This could be mediated in part by isoforms. Nuclear localization of and Ser15 phosphorylation of p53 are greater in resting muscle after high-intensity interval training239, which might help to preserve mitochondrial content and function257. By contrast, protein is unchanged after resistance training, whereas the isoform 4 () protein is preferentially enriched235. stimulates muscle hypertrophy and is associated with enhanced Igf1 expression229, insulin-like growth factor 1 (IGF1)–serine/threonine protein kinase (AKT) and mTORC1 signalling234 and downregulation of Mstn mRNA in mouse muscle229. However, unlike (ref. 280), does not coactivate oestrogen-related receptor- ()229 and has no effect on oxidative phosphorylation enzymes229,235. Appreciable overlap in the initial stages of exercise training probably underlies the degree of shared adaptation between endurance and resistance exercise (see the sections ‘Skeletal muscle responses to acute exercise’ and ‘Skeletal muscle adaptations to long-term exercise’). Depending on individual predisposition (Box 1), dedicated training of a certain exercise modality could amplify discrete differences in the adaptive response, resulting in distinct muscle adaptations and the development of specific phenotypes over time13. Evidence showing that combined endurance and resistance training can blunt muscle hypertrophy in humans is scarce184,185 but concurrent training could impede gains in explosive strength184. Still, a combined exercise regime may offer dual benefits for most individuals12. ACTRIIB, activin receptor type 2B; ECM, extracellular matrix; MFF, mitochondrial fission factor; MCU, mitochondrial calcium uniporter; NFATc1, nuclear factor of activated T cells, cytoplasmic 1; NICD, Notch intracellular domain; NRF2, nuclear factor erythroid 2-related factor 2; p38, p38 mitogen-activated protein kinase; RPS6, ribosomal protein S6; SMAD, mothers against decapentaplegic homologue; Ub, ubiquitin; , MAP3K20 isoform-.