Figure 5: OSM tumor initiating capacity requires MAPK activation.
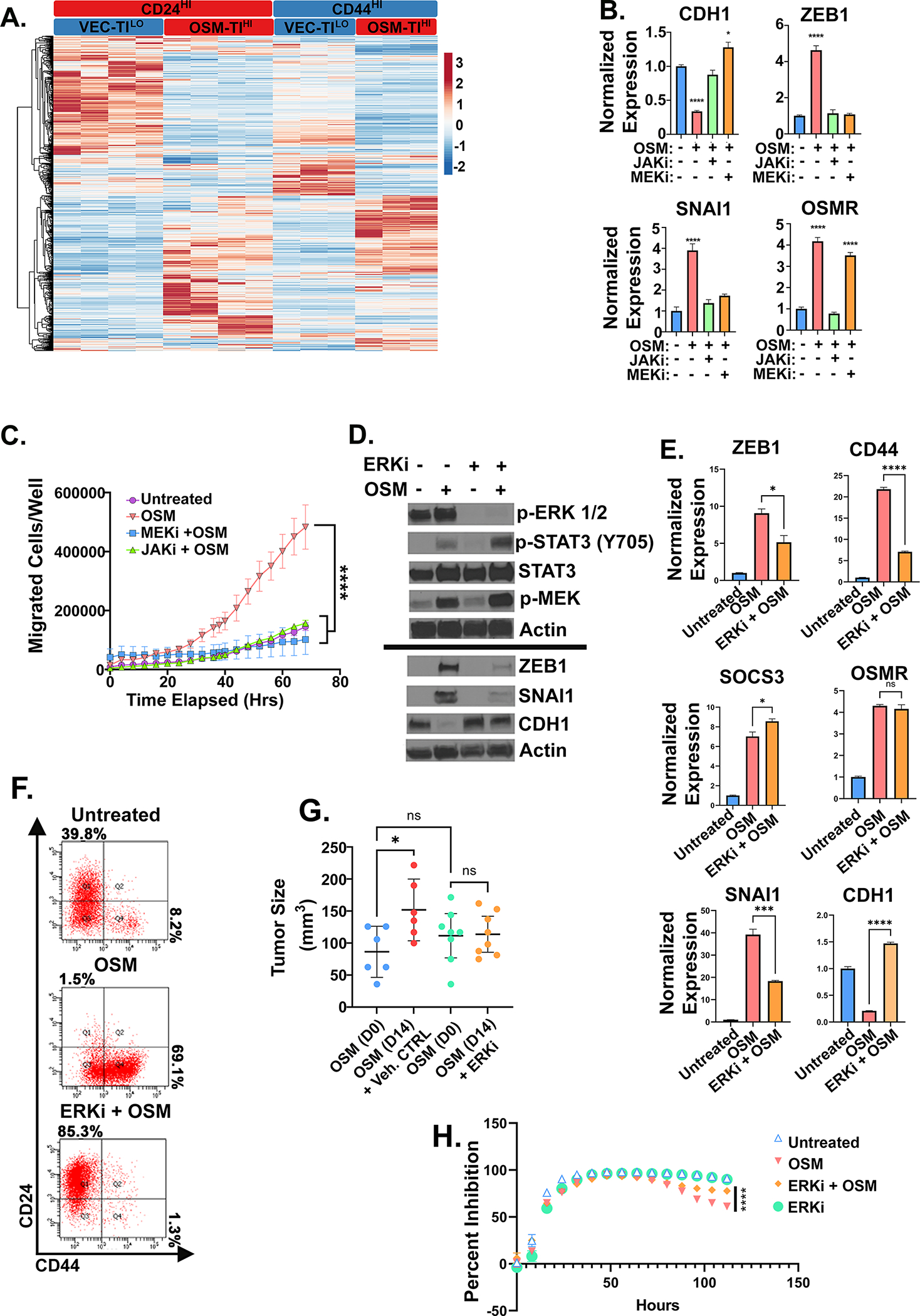
A Heatmap of RNA-seq analysis of HPAC derivatives expressing OSM (or control, VEC) used for defining the unique, stem-like program underlying OSM-induced tumor-initiation and gemcitabine resistance described in Table 1 with (n=2) for CD24HI-VEC and -OSM in a puromycin selection lentiviral plasmid, in addition to (n=2) CD24HI-VEC and –OSM and CD44HI-VEC and -OSM in a neomycin selection lentiviral plasmid. B Quantitative PCR of HPAC cells treated with non-cytotoxic/non-cytostatic doses of MEKi (U0126) or JAKi (Ruxolitinib) with or without OSM for 7 days with media changes every 48 hours. C Transwell migration of HPACs with MEKi (U0126) and JAKi, (Ruxolitinib) with or without recombinant OSM. Data is shown as mean ± S.D., and statistical significance was determined by two-way ANOVA and multiple comparisons test where ****P<0.0001. D Western blot, E quantitative PCR, and F flow cytometry of HPACs treated with rOSM with or without ERKi (SCH772984) for 5 days. E Statistical significance of quantitative PCR was determined by one-way ANOVA ns= not significant, *P<0.05, ***P<0.001, ****P<0.0001. G Tumor size measured by caliper after 1 week of tumor growth followed by 14 days of ERK inhibitor treatment. Statistical significance was determined by one-way ANOVA, where *P<0.01. H HPAC cells pre-treated with OSM with or without ERKi for 48 hours before the addition of 16 nM of gemcitabine. Data are plotted as cell number in gemcitabine at a specific timepoint divided by cell number of the corresponding untreated control at the same timepoint. Statistical significance was determined by two-way ANOVA where ****P<0.0001.
