Abstract
Introduction:
Diffusion-weighted imaging (DWI) on MRI-linear accelerator (MR-linac) systems can potentially be used for monitoring treatment response and adaptive radiotherapy in head and neck cancers (HNC) but requires extensive validation. We performed technical validation to compare six total DWI sequences on an MR-linac and MR simulator (MR sim) in patients, volunteers, and phantoms.
Methods:
Ten human papillomavirus-positive oropharyngeal cancer patients and ten healthy volunteers underwent DWI on a 1.5 T MR-linac with three DWI sequences: echo planar imaging (EPI), split acquisition of fast spin echo signals (SPLICE), and turbo spin echo (TSE). Volunteers were also imaged on a 1.5 T MR sim with three sequences: EPI, BLADE (vendor tradename), and readout segmentation of long variable echo trains (RESOLVE). Participants underwent two scan sessions per device and two repeats of each sequence per session. Repeatability and reproducibility within-subject coefficient of variation (wCV) of mean ADC were calculated for tumors and lymph nodes (patients) and parotid glands (volunteers). ADC bias, repeatability/reproducibility metrics, SNR, and geometric distortion were quantified using a phantom.
Results:
In vivo repeatability/reproducibility wCV for parotids were 5.41%/6.72%, 3.83%/8.80%, 5.66%/10.03%, 3.44%/5.70%, 5.04%/5.66%, 4.23%/7.36% for EPIMR-linac, SPLICE, TSE, EPIMR sim, BLADE, RESOLVE. Repeatability/reproducibility wCV for EPIMR-linac, SPLICE, TSE were 9.64%/10.28%, 7.84%/8.96%, 7.60%/11.68% for tumors and 7.80%/9.95%, 7.23%/8.48%, 10.82%/10.44% for nodes. All sequences except TSE had phantom ADC biases within ± 0.1×10−3 mm2/s for most vials (EPIMR-linac, SPLICE, and BLADE had 2, 3, and 1 vials out of 13 with larger biases, respectively). SNR of b = 0 images was 87.3, 180.5, 161.3, 171.0, 171.9, 130.2 for EPIMR-linac, SPLICE, TSE, EPIMR sim, BLADE, RESOLVE.
Conclusion:
MR-linac DWI sequences demonstrated near-comparable performance to MR sim sequences and warrant further clinical validation for treatment response assessment in HNC.
Keywords: Magnetic resonance imaging, Diffusion-weighted Imaging, MR-guided radiation therapy, MR-Linac, Test-retest, Apparent diffusion coefficient, Repeatability and reproducibility
Diffusion-weighted imaging (DWI) is a quantitative magnetic resonance imaging (MRI) technique that measures diffusion of water molecules in tissue, a surrogate of tissue cellularity. DWI has many applications for head and neck cancer (HNC) imaging, including lesion characterization and prediction and treatment response assessment [1–5]. Several recent studies have focused on understanding how serial DWI throughout chemotherapy and/or radiotherapy (RT) can be used to monitor response and adapt treatments based on individual response [5–12]. However, longitudinal imaging is burdensome to patients and clinicians and is generally infeasible outside of specialized research studies.
The clinical implementation of hybrid MRI/linear accelerator (MR-linac) devices has made it possible to acquire quantitative MRI sequences during every RT treatment fraction [13–17]. Current MR-linac systems enable on-line treatment plan adaptation based on changes in tumor size and shape and anatomical deformations. With further software development and validation of quantitative MRI sequences, biological image-guided adaptive RT on MR-linac systems may soon become a clinical reality [18,19].
Still, hardware modifications of current MR-linac systems to accommodate linear accelerator integration introduce additional challenges for acquiring robust quantitative MRI information. The 1.5 T MR-linac employs a split gradient coil design to allow radiation beam passage, which may contribute to magnetic field gradient non-linearities [20,21]. The maximum gradient strength and slew rate of this system are lower than conventional MRIs, which necessitates longer diffusion times for the same b-value and reduces the signal-to-noise ratio (SNR) [20]. The radiolucent 2×4 channel body coil array also reduces SNR compared to other commonly used coils [22]. In light of these challenges, the MR-Linac Consortium has released guidelines for acquiring DWI on this system [20], which has informed the selection of sequence parameters in this study.
In RT, spatial accuracy of images is crucial to ensure the precise delivery of radiation. Single-shot echo planar imaging (EPI), the most commonly used readout method for DWI, is prone to severe geometric distortions and susceptibility artifacts, especially in the head and neck [23]. Turbo spin echo (TSE)-based DWI sequences have been shown to improve spatial fidelity [23,24] and are of interest for biological image-guided adaptive RT applications. However, destructive interference between spin echoes and stimulated echoes can reduce SNR in TSE-DWI. An alternative TSE-based method, “split acquisition of fast spin echo signals” (SPLICE), acquires the spin echo and stimulated echo contributions separately to preserve SNR while maintaining the spatial accuracy of TSE [25,26].
In this study, we investigate the performance of EPI, TSE, and SPLICE DWI sequences on the 1.5 T MR-linac and compare them to three DWI sequences on a 1.5 T diagnostic-quality MR simulation (MR sim) scanner. The MR sim sequences include EPI and two additional low-distortion sequences: “BLADE” (vendor tradename), which is a hybrid TSE/gradient echo acquisition that uses a radial blade k-space trajectory [27,28], and “readout segmentation of long variable echo trains” (RESOLVE), a multi-shot EPI sequence [29,30]. In this R-IDEAL stage 2a1 study [31], we perform technical validation of these six DWI sequences using data from human papillomavirus-positive (HPV + ) oropharyngeal cancer patients, healthy volunteers, and a diffusion phantom.
Methods
Participants and imaging
Ten patients and ten healthy volunteers were included in this study. All participants provided written informed consent; patients were consented to the MOMENTUM observational clinical trial (NCT04075305) [32] and volunteers to an internal volunteer imaging protocol, both approved by MD Anderson Cancer Center’s institutional review board. Inclusion criteria for patients included non-recurrent, histologically confirmed HPV + oropharyngeal cancer with no prior history of cancer therapy. All imaging occurred between diagnosis and the start of treatment. Clinical demographics are in Table 1.
Table 1.
Clinical demographics for patients and healthy volunteers. All patients had non-recurrent, histologically confirmed HPV + oropharyngeal cancer.
| Characteristic | Patients | Healthy Volunteers |
|---|---|---|
| Sex | ||
| Male | 10 (100%) | 7 (70%) |
| Female | 0 (0%) | 3 (30%) |
| Age (years) | ||
| Median (range) | 61 (58–74) | 30 (24–43) |
| Disease sub-site | ||
| Tonsil | 4 (40%) | |
| Base of tongue | 6 (60%) | |
| T stage | ||
| T1 | 5 (50%) | |
| T2 | 2 (20%) | |
| T3 | 3 (30%) | |
| N stage | ||
| N1 | 7 (70%) | |
| N2 | 3 (30%) | |
| M stage | ||
| M0 | 10 (100%) | |
| Clinical Stage | ||
| I | 6 (60%) | |
| II | 4 (40%) |
Patients and volunteers were imaged on a 1.5 T MR-linac (Unity; Elekta AB; Stockholm, Sweden) with a 3-D fat-suppressed T2-weighted MRI sequence and three DWI sequences: EPIMR-linac, SPLICE, and TSE. Volunteers were also imaged on a 1.5 T MR sim (MAGNETOM Aera; Siemens Healthcare; Erlangen, Germany) with a multi-slice fat-suppressed T2-weighted MRI sequence and three DWI sequences: EPIMR sim, BLADE, and RESOLVE. Sequence descriptions and parameters are shown in Supplementary Tables S1 and S2. All in vivo images were acquired in the axial slice orientation. Diffusion gradients were applied along three orthogonal directions, and the images were combined into a single trace-weighted image. The MR-linac acquisitions used a rigid radiolucent 2×4 channel array coil [33], and the MR sim acquisitions used two 4-channel flex coils and integrated table coils. Acquisition times (minutes) were 3.07, 7.38, 4.90, 2.93, 7.13, and 6.75 for EPIMR-linac, SPLICE, TSE, EPIMR sim, BLADE, and RESOLVE, respectively.
The diffusion b-values used were 0, 150, 500 s/mm2 for the MR-linac, 0, 500 s/mm2 for EPI on the MR sim, and 0, 800 s/mm2 for BLADE and RESOLVE. The choice of b-values for the MR sim was based on scan protocols used in clinical trials at our institution, which were used in this study without modification. MR-linac b-values were chosen based on the MR-Linac Consortium’s recommendations [20]. However, because only two b-values were used for the MR sim images, ADC maps for the MR-linac images were reconstructed only with the 0 and 500 s/mm2 images for direct comparison to the MR sim. Although different high b-values were used for BLADE and RESOLVE, b-value linearity data shown in the Supplementary Data (Figure S1) demonstrates that there is no significant b-value dependence for these sequences. ADC maps were reconstructed using the built-in software on each scanner. An analysis in the Supplementary Data explores differences in ADC values and repeatability/reproducibility metrics between ADC maps reconstructed with b = 0,500 mm2/s (ADC0,500) and b = 150,500 mm2/s (ADC150,500) to investigate whether the perfusion contributions to the signal at low b-values impact the ADC precision.
Each study participant underwent two scan sessions per device. The first and second time points occurred at least one day apart, depending on clinical scheduling availability; mean (range) number of days between scans was 8 (1–15) for patients and 6 (1–21) for volunteers. All participants were imaged in custom RT immobilization masks to minimize motion and ensure setup reproducibility. During each session, participants were scanned twice with each DWI sequence, with a short “coffee break” out of the mask between each set to test repeatability.
In vivo data analysis
A radiologist with 5 years of experience delineated the primary tumor and pathological lymph nodes (patients) and parotid glands (volunteers). One patient did not have an MR-visible primary tumor. A total of 9 primary tumors, 30 lymph nodes, and 20 parotid glands were analyzed. Regions of interest were delineated on T2-weighted images and rigidly copied to the high-b-value image of each DWI then manually edited to account for any distortion. Segmentations were rigidly copied to corresponding ADC maps.
Repeatability/reproducibility metrics (within-subject coefficient of variation (wCV) of mean ADC) were calculated for each DWI sequence and structure type according to the Quantitative Imaging Biomarker Alliance (QIBA) consensus recommendations [34,35]. 95% confidence intervals for wCV were calculated using a chi-square statistic with n(K-1) degrees of freedom, where n is the number of sets of replicate measurements and K is the number of repeats [36]. For the repeatability (i.e. short-term) wCV calculation, there were two pairs of replicate measurements per patient/volunteer (two replicate images from the first scan session and two replicate images from the second scan session). For the reproducibility (i.e. long-term) wCV calculation, there were also two pairs of replicate measurements per patient/volunteer (the first images from the first and second scan sessions were paired, and the second images from the first and second scan sessions were paired). Thus, the total number of data points used for analysis were 18 primary tumors, 60 lymph nodes, and 60 parotid glands.
Bland-Altman analysis was performed between all pairs of DWI sequences to measure differences in calculated ADC values. Values from all four imaging time points were included. Mean difference (bias) values, 95% confidence intervals for the mean differences, and 95% Bland-Altman limits of agreement were calculated in JMP (v15.0.0; SAS Institute Inc.; Cary, NC, USA) using the Method Comparison add-in. Bland-Altman analysis was also performed to assess differences in ADC values between ADC0,500 and ADC150,500 maps (Supplementary Data).
Phantom data acquisition and analysis
Four sequential repeats of each DWI sequence were acquired of the QIBA diffusion phantom (model 128; CaliberMRI; Boulder, CO) for ADC bias, repeatability wCV and repeatability coefficient (RC), and SNR calculations. Images were acquired in the coronal slice orientation. The phantom was held constant at 0 °C using an ice water bath, and the temperature was verified before and after each scan session. ADC bias, wCV, RC, and SNR were calculated for each sequence using methods described in the QIBA guidelines [34,35]. ADC bias was calculated for each vial by measuring the mean ADC value in each region of interest (ROI) and subtracting the manufacturer-provided ADC. The SNR calculation involved creating a “signal image” from the voxel-wise average of the four images and a “temporal noise image” from the voxel-wise standard deviation. SNR is equal to the ROI mean for the “signal image” divided by the ROI mean for the “temporal noise image.” A single repeat of each sequence was also acquired at a separate time point for reproducibility measurements, i.e. wCV and reproducibility coefficient (RDC). ADC bias was calculated for each vial, while the other quantities were calculated only in the central phantom vial, per the guidelines.
Geometric distortion of each sequence was assessed using the grid section of the American College of Radiology MRI phantom and by reducing the in-plane resolution to 1.5 mm, which is described in the Supplementary Data.
Results
Representative images of the six DWI sequences of a patient and volunteer are shown in Fig. 1. In vivo mean ADC values and repeatability/reproducibility wCV values are shown in Table 2. Reproducibility wCV values were higher than repeatability wCV values for all sequences and structure types. For the MR-linac sequences with which both patients and volunteers were imaged, both repeatability and reproducibility wCV values were consistently higher for tumors and nodes than for parotid glands. In parotid glands, wCV values were slightly higher overall for the MR-linac sequences compared to the MR sim sequences.
Fig. 1.
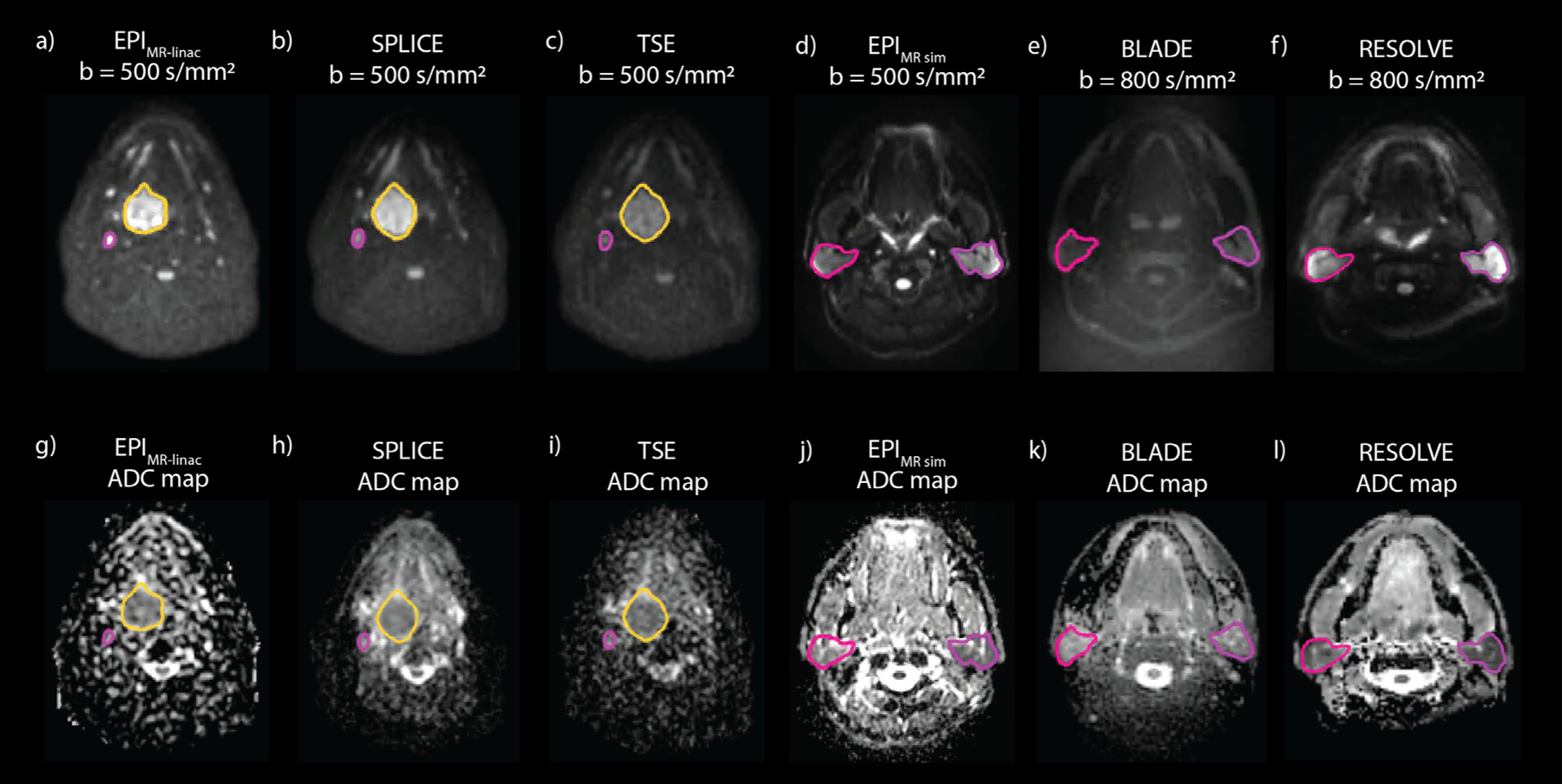
Representative high b-value images and ADC maps of a patient imaged with EPI (a, g), SPLICE (b, h), and TSE (c, i) on the MR-linac and a volunteer imaged with EPI (d, j), BLADE (e, k), and RESOLVE (f, l) on the MR sim. The primary tumor (yellow) and a lymph node (pink) are segmented on the patient images, while the parotid glands are segmented on the volunteer images.
Table 2.
In vivo mean ADC values and repeatability/reproducibility wCV for each DWI sequence. Volume and mean ADC values are represented as mean ± standard deviation. wCV values are represented as wCV (upper and lower 95% confidence interval) and expressed as a percentage. Sequences are color-coded to group sequences with similar acquisition mechanisms that can be directly compared.
| EPIMR-linac | SPLICE | TSE | EPIMR sim | BLADE | RESOLVE | ||
|---|---|---|---|---|---|---|---|
| Sequence Description | Single-shot SE EPI | Single-shot TSE with split acquisition of spin echo and stimulated echo | Single-shot TSE | Single-shot SE EPI | Multi-shot radial TGSE | Multi-shot SE EPI | |
| Parotid Glands Volume (cm3): 32.9 ± 16.0 | Mean ADC (×10−3 mm2/s) | 1.14 ± 0.17 | 1.20 ± 0.23 | 0.93 ± 0.18 | 1.49 ± 0.20 | 1.14 ± 0.15 | 1.19 ± 0.16 |
| Repeatability wCV (%) | 5.41 (4.42 – 6.97) | 3.83 (3.13 – 4.94) | 5.66 (4.63 – 7.29) | 3.44 (2.81 – 4.43) | 5.04 (4.12 – 6.50) | 4.23 (3.46 – 5.45) | |
| Reproducibility wCV (%) | 6.72 (5.49 – 8.66) | 8.80 (7.19 – 11.34) | 10.03 (8.20 – 12.92) | 5.70 (4.64 – 7.40) | 5.66 (4.62 – 7.29) | 7.36 (5.99 – 9.56) | |
| Primary Tumors Volume (cm3): 10.7 ± 10.4 | Mean ADC (×10−3 mm2/s) | 1.33 ± 0.22 | 1.80 ± 0.19 | 1.36 ± 0.19 | |||
| Repeatability wCV (%) | 9.64 (7.23 – 14.45) | 7.84 (5.84 – 11.93) | 7.60 (5.70 – 11.40) | ||||
| Reproducibility wCV (%) | 10.28 (7.65 –15.64) | 8.96 (6.56 – 14.14) | 11.68 (8.70 – 17.78) | ||||
| Lymph Nodes Volume (cm3): 7.3 ± 8.0 | Mean ADC (×10−3 mm2/s) | 1.38 ± 0.40 | 1.53 ± 0.36 | 1.31 ± 0.42 | |||
| Repeatability wCV (%) | 7.80 (6.59 – 9.55) | 7.23 (6.06 – 8.96) | 10.82 (9.15 – 13.25) | ||||
| Reproducibility wCV (%) | 9.95 (8.37 – 12.25) | 8.48 (7.00 – 10.78) | 10.44 (8.79 – 12.86) |
Differences in mean ADC and wCV between ADC0,500 and ADC150,500 maps are shown in Supplementary Table S3. Mean ADC values were consistently higher for ADC0,500 than for ADC150,500 maps. Both repeatability and reproducibility wCV values were higher for the ADC150,500 maps in nearly all cases.
Bland-Altman analysis (Fig. 2, Supplementary Figure S2) revealed statistically significant biases for all pairs of DWI sequences except BLADE-EPIMR-linac and RESOLVE-SPLICE. Mean differences between sequences ranged from 0.00 (BLADE-EPIMR-linac) to 0.56×10−3 mm2/s (EPIMR sim-TSE). For the MR-linac sequences where both patient and volunteer data were compared, the SPLICE-EPIMR-linac and TSE-EPIMR-linac combinations showed different biases among structure types.
Fig. 2.
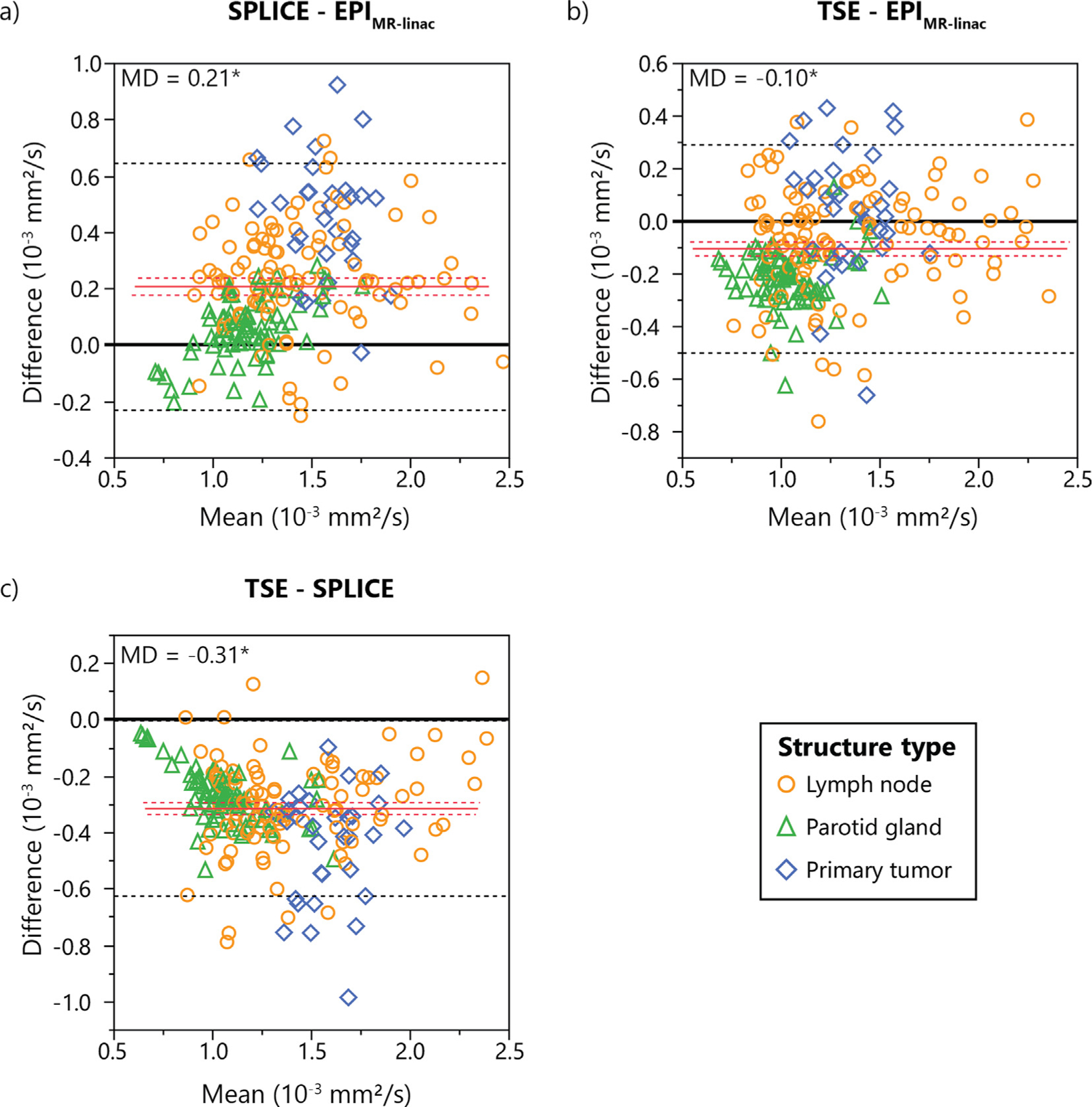
Bland-Altman plots showing differences in measured ADC values between each pair of the three MR-linac DWI sequences. Mean differences (MD) are represented by solid red lines with 95% confidence intervals of the MD represented by dotted red lines. 95% limits of agreement are represented by dotted black lines. Statistically significant biases are represented by * (p < 0.05).
Bland-Altman plots showing the differences in calculated ADC values between ADC0,500 and ADC150,500 maps are shown in Supplementary Figure S3. ADC0,500 overestimated ADC150,500 by 0.34, 0.48, and 0.31 ×10−3 mm2/s for EPIMR-linac, SPLICE, and TSE, respectively. Phantom ADC bias results in all phantom vials are shown in Fig. 3. For all MR sim sequences, the range of ADC bias values fell within ± 0.1×10−3 mm2/s (except BLADE in the vials with true ADC value of 1.127×10−3 mm2/s). For the MR-linac sequences, EPIMR-linac and SPLICE had all ADC bias values fall within ± 0.1×10−3 mm2/s for the first four vials, but ADC overestimation occurred for EPIMR-linac and SPLICE at the lowest two ADC values. TSE underestimated ADC by more than 0.1 mm2/s for nearly all vials. In general, the MR sim sequences were more precise than the MR-linac sequences. ADC values for the MR-linac sequences tend to be consistent for each vial across replicate images but inconsistent across different vials with the same true ADC value. (For clarity, a similar graph is shown in Supplementary Figure S4 showing the phantom ADC bias results combining data points from all vials with the same true ADC value.).
Fig. 3.
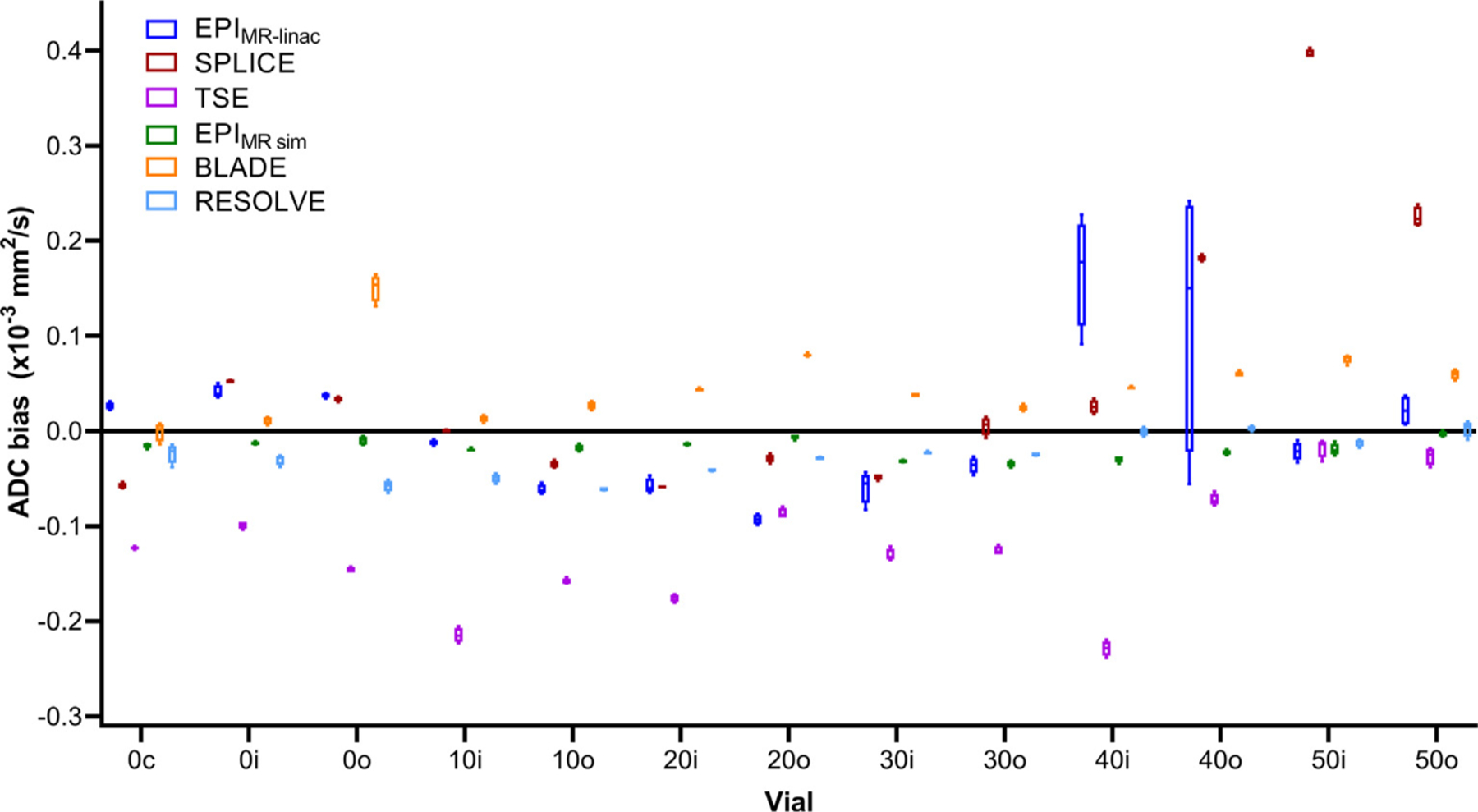
Phantom ADC bias results from four replicate image acquisitions for each individual phantom vial. Each data point within each box-and-whisker plot represents the difference between the mean ADC value in an ROI and the true ADC value for each phantom vial. The numbers in the phantom vial names refer to the concentrations of polymer, which correspond to the manufacturer-provided true ADC values (1.127×10−3 mm2/s for 0, 0.843×10−3 mm2/s for 10, 0.607×10−3 mm2/s for 20, 0.403×10−3 mm2/s for 30, 0.248×10−3 mm2/s for 40, and 0.128×10−3 mm2/s for 50). The letters in the vial names refer to the spatial location of the vial within the phantom (“c” for “center,” “i” for “inner,” and “o” for “outer”).
Phantom ADC bias (in the central vial), %CV, repeatability and reproducibility metrics, and SNR are shown in Table 3. Tolerance values from the QIBA Profile [35] are also included for reference. All sequences met the ± 0.040×10−3 mm2/s criterion for ADC bias except SPLICE and TSE (−0.057 and −0.123×10−3 mm2/s, respectively). For %CV, only EPIMR sim and BLADE met the 2% threshold, but all MR-linac sequences were close (≤3.62%). RESOLVE had a much higher %CV (8.30%). For RC, all MR-linac sequences and EPIMR sim fell under 0.0150×10−3 mm2/s, but BLADE and RESOLVE did not (0.0269 and 0.0279 ×10−3 mm2/s, respectively). All sequences were within the RDC limit. All sequences exceeded the SNR threshold of 50, but EPI had the lowest SNR (87.3 for b = 0).
Table 3.
Phantom ADC bias, repeatability metrics, and SNR for each DWI sequence. ADC bias was calculated using the central phantom vial. The ADC bias and within-ROI %CV are expressed as the mean ± standard deviation of the values measured in each of the four replicate images. Tolerance values from the QIBA Profile are included in the last column for comparison.
| EPI (MR-linac) | SPLICE (MR-linac) | TSE (MR-linac) | EPI (MR sim) | BLADE (MR sim) | RESOLVE (MR sim) | QIBA tolerance values | |
|---|---|---|---|---|---|---|---|
| ADC bias (10−3 mm2/s) | 0.026 ± 0.004 | −0.057 ± 0.003 | −0.123 ± 0.002 | −0.015 ± 0.002 | −0.001 ± 0.010 | −0.024 ± 0.010 | ≤ ±0.040 |
| %CV (within-ROI) | 2.55 ± 0.71 | 3.14 ± 0.11 | 3.62 ± 0.30 | 1.76 ± 0.12 | 1.85 ± 0.12 | 8.30 ± 3.04 | ≤ 2 |
| RC (×10−3 mm2/s) | 0.0107 | 0.0082 | 0.0041 | 0.0067 | 0.0269 | 0.0279 | ≤ 0.0150 |
| Repeatability wCV (%) | 0.33 | 0.28 | 0.15 | 0.22 | 0.86 | 0.91 | N/A |
| RDC (×10−3 mm2/s) | 0.0305 | 0.0254 | 0.0027 | 0.0055 | 0.0416 | 0.0049 | ≤ 0.0650 |
| Reproducibility wCV (%) | 0.96 | 0.85 | 0.10 | 0.18 | 1.36 | 0.16 | N/A |
| SNR b = 0 s/mm2 | 87.3 (78.6 – 96.1) |
180.5 (168.0 – 193.0) |
161.3 (146.7 – 175.9) |
171.0 (160.8 – 181.2) |
171.9 (147.5 – 196.3) |
130.2 (112.3 – 148.0) |
≥ 50 ± 5 |
| SNR b = 150 s/mm2 | 118.9 (102.8 – 135.0) |
193.1 (183.6 – 202.6) |
266.2 (242.2 – 290.1) |
N/A | N/A | N/A | N/A |
| SNR b = 500 s/mm2 or b = 800 s/mm2 | 106.7 (94.6 – 118.8) (b = 500) |
184.0 (174.8 – 193.3) (b = 500) |
261.6 (241.6 – 281.7) (b = 500) |
232.2 (222.8 – 241.7) (b = 500) |
104.1 (89.4 – 118.7) (b = 800) |
97.3 (83.5 – 111.1) (b = 800) |
N/A |
The geometric distortion analysis (Supplementary Figure S7) showed that all sequences had single-point distortions less than the limiting resolution of 1.5 mm in the frequency-encoding direction. In the phase-encoding direction, EPIMR-linac showed the greatest distortion, with values as high as 11.0 mm. EPIMR sim and RESOLVE showed small amounts of distortion up to 2.3 mm and 1.6 mm, respectively.
Discussion
The goals of this study were 1) to compare the performance of DWI on the 1.5 T MR-linac with a 1.5 T diagnostic quality MR sim and 2) to select an optimal DWI sequence for HNC on the MR-linac. To accomplish these goals, we quantified the ADC repeatability and reproducibility, ADC bias, and SNR of three DWI sequences each on a 1.5 T MR-linac and a 1.5 T MR sim in vivo and in a phantom.
ADC repeatability was previously quantified for various HNCs on diagnostic MRI systems by Paudyal et al. [37]. They measured a wCV of 2.38% for lymph nodes in a mixed cohort of 9 HNC patients imaged with EPI on a 3 T MRI. This value is substantially lower than the wCV values measured for lymph nodes with the three MR-linac sequences in our study but is also lower than the values measured for parotid glands on the MR sim, which may be attributable to differences in the MRI hardware (field strength, gradients, and coil hardware) as well as the image acquisition and post-processing techniques (b-values, ADC fitting model, etc.).
Repeatability/reproducibility of quantitative imaging biomarkers have been previously quantified on the MR-linac in multiple disease sites. Habrich et al. [38] measured the reproducibility of ADC in a mixed cohort of HNC patients using a single-shot EPI sequence with b-values of 150 and 500 s/mm2 and calculated RC values of 13.3% and 15.2% for left and right parotid glands, respectively, 31.3% for primary tumors, and 23.5% for lymph nodes. Per QIBA guidelines, RC is calculated by multiplying the repeatability wCV by 2.77 [34]; our RC values are 15.0%, 26.7%, and 21.6% for parotid glands, primary tumors, and lymph nodes. Our results are similar despite a few differences in study design: their test–retest scans were performed throughout the course of RT instead of pretreatment, and patients were not repositioned between scans. Next, Lawrence et al. [16] measured the ADC repeatability and reproducibility of brain tumors and healthy tissues on the MR-linac and a 1.5 T diagnostic scanner. wCV values on the MR-linac were within 5% and were comparable to the diagnostic system. Kooreman et al. [15] assessed the reproducibility of intravoxel incoherent motion parameters in prostate cancer patients on the MR-linac and found the RDC of the diffusion coefficient to be 0.09×10−3 mm2/s for non-cancerous prostate and 0.44×10−3 mm2/s for tumors. For a HNC cohort on a 0.35 T MR-linac, Yang et al. [17] did not explicitly calculate reproducibility metrics but found the ADC of the brainstem to be within 0.47–0.57×10−3 mm2/s across repeat imaging sessions. These studies demonstrate the robustness of quantitative imaging biomarkers on the MR-linac, suggesting their potential for longitudinal quantitative imaging and biological image-guided adaptive treatments. However, extensive validation of quantitative imaging biomarkers and standardization of scan protocols across sites is necessary for large-cohort, multi-center studies [13,19].
The QIBA Diffusion Profile [35] provides acceptability criteria for phantom metrics, which are included in Table 3 for comparison to measured values. All sequences except EPIMR sim violated at least one tolerance value. However, it is important to note that these criteria are defined for system performance evaluation using an EPI sequence with specific parameters. Thus, they are not directly applicable to evaluate the sequences used in this paper but may serve as starting points for acceptability criteria based on clinical needs. In particular, the lowest ADC components of the phantom are best characterized using b = 2000 s/mm2; use of lower maximum b-values can result in higher wCVs [39].
Our phantom data showed that in MR-linac sequences only, ADC values varied substantially across different vials with the same diffusivity but not across replicate images within the same vial, suggesting that spatial inhomogeneities are more significant for the MR-linac than the MR sim. This may be attributed to gradient non-linearities: a gradient non-linearity correction is automatically included in the vendor-provided ADC reconstruction for the Siemens MR sim, but the Philips gradient non-linearity feature was not available on the Elekta/Philips MR-linac at the time of writing. Kooreman et al. [20] found similar results with a homogeneous diffusion phantom imaged on an MR-linac and MR sim but found that the spatial inhomogeneities persisted on the MR-linac data even after applying an off-line correction for gradient non-linearities. They attributed the differences to the split gradient coil design on the MR-linac, which is necessary to accommodate the radiation treatment beam but induces eddy currents closer to isocenter, causing magnetic field inhomogeneities.
Parotid gland wCV values were higher overall for the MR-linac compared to the MR sim, but differences were small (within 2.22% and 4.37% for repeatability and reproducibility, respectively). However, to determine whether the precision of the MR-linac DWI sequences is within a clinically acceptable range for longitudinal treatment response evaluation, QIBA recommends calculating the threshold change in ADC that can be attributed to a true biological change rather than measurement noise within 95% confidence by multiplying the reproducibility wCV values by 2.77 [34]. For the three MR-linac sequences, these values range from 18.6% – 27.8% for parotid glands, 24.8% – 32.4% for primary tumors, and 23.5% – 28.9% for lymph nodes. Several studies have analyzed the differences in ADC from baseline to mid-RT between groups of patients with different response to treatment, which have fallen within these ranges. Matoba et al. found that a threshold of 24% change in ADC between baseline and the third week of RT for primary tumors could predict progression-free survival in a mixed cohort of HNC patients, while ADC changes in lymph nodes were not significantly different between the two groups [40]. Similarly, Khattab et al. found an optimal threshold of 33% change in ADC between baseline and mid-RT in primary tumors to predict local failure after chemo-RT, also in a mixed cohort of HNC patients [41]. These studies cannot be directly compared to our results due to the differences in DWI acquisition parameters and patient populations, but this comparison suggests that in vivo DWI of HNC is possible on the 1.5 T MR-linac with acceptable repeatability/reproducibil ity, laying the foundation for future clinical studies.
Comparing the MR-linac sequences, EPIMR-linac and SPLICE had more accurate phantom ADC values than TSE, with the exception of SPLICE at low ADC values. For repeatability, SPLICE had the lowest overall wCV values and highest overall SNR. Both SPLICE and TSE had undetectable levels of geometric distortion, while EPIMR-linac had the highest distortion of all six sequences. Based on these data and our clinical preference for a low-distortion DWI sequence, SPLICE is the optimal sequence for HNC imaging on the MR-linac. However, a disadvantage of SPLICE is the long acquisition time (7 minutes for 3b-values).
For the Bland-Altman analysis with the in vivo MR-linac data, different mean ADC biases were observed for different structure types (Fig. 2). Both SPLICE and TSE compared to EPIMR-linac demonstrated the highest ADC biases for primary tumors and the lowest for parotid glands, but there were no differences among structure types between SPLICE and TSE. Though the exact reason for these differences is not well understood, they are likely caused by a combination of the different anatomical composition of each structure type (i.e. the degree of restricted diffusion vs. unrestricted diffusion vs. perfusion) and the different signal acquisition mechanisms of EPI and the two TSE-based sequences (rapid gradient switching vs. rapid refocusing pulses). Although both primary tumors and parotid glands have a high degree of perfusion, tumors typically have more perfusion due to the higher density and permeability of the tumor vasculature compared to normal capillary networks [42]. Further, blood in tumor capillaries tends to have more deoxygenated blood than in healthy capillaries [43], and the paramagnetic deoxygenated hemoglobin introduces T2* signal decay [44]. EPI sequences are more susceptible to T2* decay than TSE-based sequences due to the lack of refocusing pulses [44], so the difference in the perfusion signal contribution between TSE-based sequences and EPI would be greater for the primary tumors than for the parotid glands, causing the differences seen in Fig. 2. Another potential contributing factor to these differences may be magnetization transfer effects caused by the repeated refocusing pulses in the TSE-based sequences, which may also affect the image signal and ADC values relative to the EPI [45]. Finally, differences in SNR between sequences or the potential presence of artifacts could cause differences in ADC between sequences, although further investigation is required to truly understand this phenomenon.
One surprising result is that the SNR of EPIMR-linac was much lower than for SPLICE and TSE. We used the consensus EPI protocol that had been distributed among MR-Linac Consortium [46] sites without modification for comparison across sites. Because no consensus SPLICE or TSE protocols existed, these sequences were optimized in-house based on both phantom and in vivo experiments, with analyses of SNR, contrast-to-noise ratio, ADC accuracy, and qualitative assessment by expert readers [47]. Thus, different sequence parameters were used between EPI and SPLICE/TSE that could impact the SNR. First, the diffusion time was much longer in EPIMR-linac (47.4 ms) than in SPLICE (32.8 ms) or TSE (31.4 ms), which results in more T2 decay and reduces SNR. A second potential explanation is that short inversion time inversion recovery (STIR) was used for fat suppression for EPIMR-linac and spectral presaturation with inversion recovery (SPIR) for SPLICE and TSE. STIR uses inversion recovery to nullify fat signal but reduces signal from all tissues, resulting in reduced SNR. SPIR uses a spectrally selective inversion pulse to improve SNR from non-fat tissues [48]. We chose SPIR over STIR for the SPLICE and TSE to maximize SNR. If SPIR is used for EPI, SNR and reproducibility would likely improve. However, poor fat suppression in EPI causes large chemical shift artifacts, while the non-suppressed fat in TSE and SPLICE sequences only shows a small chemical shift.
One limitation of this study was our relatively small sample size of 10 patients and 10 volunteers, with a total of 9 primary tumors, 30 lymph nodes, and 20 parotid glands for analysis. According to the QIBA recommendations [34] based on the work of Obuchowski and Bullen [49], a minimum of 35 subjects are required to estimate repeatability and reproducibility for tracking longitudinal changes in a quantitative imaging biomarker for treatment response. However, multiple lesions/parotid glands within the same patient can be treated as independent samples for repeatability/reproducibility calculations. We also acquired a total of four repeats of each DWI sequence over two imaging sessions per device, resulting in two pairs of replicate measurements for both the repeatability and reproducibility calculations (i.e. effectively doubling our sample size). While these cannot be considered completely independent samples, we believe that our sample sizes are large enough to demonstrate differences between the DWI sequences studied in this paper. Still, a larger number of samples, especially for primary tumors, would be required to calculate the true repeatability/reproducibility coefficients for treatment response assessment.
Another limitation was that clinical scheduling constraints prevented patients from undergoing two scans each on both the MR-linac and MR sim between the time of simulation and the start of treatment. While patients would provide the most ideal comparison between the devices, healthy volunteers were included to assess differences between the systems in parotid glands. However, our data reveal that the repeatability and ADC bias behavior of parotid glands differs from that of tumors and nodes, so future investigations using patients on both the MR-linac and MR sim should confirm the findings in this study. Also, for the experiments in the main body of the paper, the ADC maps were calculated using a low b-value of 0 s/mm2, so the signal in the in vivo images includes contributions from perfusion. The perfusion effects can be minimized by using a higher low b-value such as 150 s/mm2 [50], as was done with the MR-linac images in the Supplementary Data. Finally, the most ideal comparison between the MR-linac and a diagnostic-quality scanner would be to use a 1.5 T Philips MRI with the same DWI sequences, but we were limited by the MR sim and sequences available at our institution. Nonetheless, our goal was to evaluate whether the reduced gradient performance of the MR-linac substantially degraded the repeatability and reproducibility of the ADC values obtained compared to an MR sim; given that the Siemens Aera has similar gradient specifications compared to the Philips 1.5 T Ingenia, our inclusion of data from this device in our study serves the purpose of that comparison.
Conclusion
We have assessed the repeatability/reproducibility, ADC bias, and SNR of DWI sequences on a 1.5 T MR-linac and MR sim for HNC both in vivo and in phantoms and demonstrated near-comparable performance between the MR-linac and MR sim. These results show that the MR-linac DWI sequences are robust and worthy of further evaluation as a quantitative method of assessing treatment response in HNC.
Supplementary Material
Acknowledgements
The authors would like to acknowledge the following funding sources: B.A. McDonald has The authors would like to acknowledge the following funding sources: B.A. McDonald has received research and training support through a Ruth L. Kirschstein fellowship from NIH/NIDCR (1F31DE029093), a Dr. John J. Kopchick Fellowship from The University of Texas MD Anderson Cancer Center UTHealth Graduate School of Biomedical Sciences, and an Image-Guided Cancer Therapy T32 Fellowship from MD Anderson Cancer Center and NIH/National Cancer Institute. C.D. Fuller and J. Christodouleas receive research support from an academic-industrial partnership R01 grant from the National Institutes of Health (NIH)/National Institute of Dental and Craniofacial Research (NIDCR) (R01DE028290) directly related to this project. C.D. Fuller has received industry research support from Elekta AB. C.D. Fuller reports additional grants from NIH/National Cancer Institute (R01CA218148, 1R01CA225190, 1R01CA214825, P30CA016672, P50CA097007–10), NIH/NIDCR (1R01DE025248/R56DE025248), NIH/National Institute of Biomedical Imaging and Bioengineering (R25EB025787), Patient-centered Outcomes Research Institute (PCS-1609–36195), Sister Institute Network Fund (MD Anderson), and National Science Foundation Division of Civil, Mechanical, and Manufacturing Innovation (CMMI 1933369), outside of the scope of the current project. T.C. Salzillo is supported by The University of Texas Health Science Center at Houston Center for Clinical and Translational Sciences TL1 Program (TL1TR003169)
Footnotes
Conflict of interest statement
J. Christodouleas and A. Dresner are employees of Elekta AB and Philips, respectively, and have collaborated on this project as part of an academic-industrial partnership R01 grant (R01DE028290). C. D. Fuller has received industry research support, travel funding, and speaking honoraria from Elekta AB unrelated to the current project. M. Philippens reports institutional collaborations including financial and technical support under research agreements as well sponsorship for travel and scientific symposia from Elekta AB (Stockholm, Sweden) and Philips NV (Best, The Netherlands). D. Thorwarth reports institutional collaborations including financial and non-financial support by Elekta, Philips, Kaiku, Therapanacea and PTW Freiburg.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.radonc.2023.109717.
R-IDEAL is an evaluation framework for radiation oncology technological advancements. Stage 2a is the “development” phase, including “technical improvements, feasibility, and safety.”.
References
- [1].Payabvash S Quantitative diffusion magnetic resonance imaging in head and neck tumors. Quant Imaging Med Surg 2018;8:1052–65. 10.21037/qims.2018.10.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Salzillo TC, Taku N, Wahid KA, McDonald BA, Wang J, van Dijk LV, et al. Advances in imaging for HPV-related oropharyngeal cancer: applications to radiation oncology. Semin Radiat Oncol 2021;31:371–88. 10.1016/j.semradonc.2021.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Driessen JP, Van Kempen PMW, Van Der Heijden GJ, Philippens MEP, Pameijer FA, Stegeman I, et al. Diffusion-weighted imaging in head and neck squamous cell carcinomas: a systematic review. Head Neck 2015. 10.1002/hed.23575. [DOI] [PubMed]
- [4].Connolly M, Srinivasan A. Diffusion-weighted imaging in head and neck cancer: technique, limitations, and applications. Magn Reson Imaging Clin N Am 2018;26:121–33. 10.1016/j.mric.2017.08.011. [DOI] [PubMed] [Google Scholar]
- [5].Chang Z, Wang C. Treatment assessment of radiotherapy using MR functional quantitative imaging. World J Radiol 2015;7:1–6. 10.4329/wjr.v7.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Vandecaveye V, Dirix P, De Keyzer F, Op De Beeck K, Vander Poorten V, Hauben E, et al. Diffusion-weighted magnetic resonance imaging early after chemoradiotherapy to monitor treatment response in head-and-neck squamous cell carcinoma. Int J Radiat Oncol Biol Phys 2012;82:1098–107. 10.1016/j.ijrobp.2011.02.044. [DOI] [PubMed] [Google Scholar]
- [7].Vandecaveye V, Dirix P, De Keyzer F, Op De Beeck K, Vander V, Roebben PI, et al. Predictive value of diffusion-weighted magnetic resonance imaging during chemoradiotherapy for head and neck squamous cell carcinoma. Eur Radiol 2010;20:1703–14. 10.1007/s00330-010-1734-6. [DOI] [PubMed] [Google Scholar]
- [8].Hong J, Yao Y, Zhang Y, Tang T, Zhang H, Bao D, et al. Value of magnetic resonance diffusion-weighted imaging for the prediction of radiosensitivity in nasopharyngeal carcinoma. Otolaryngol - Head Neck Surg 2013;149:707–13. 10.1177/0194599813496537. [DOI] [PubMed] [Google Scholar]
- [9].van der Heide UA, Houweling AC, Groenendaal G, Beets-Tan RGH, Lambin P. Functional MRI for radiotherapy dose painting. Magn Reson Imaging 2012;30:1216–23. 10.1016/j.mri.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Houweling AC, Wolf AL, Vogel WV, Hamming-Vrieze O, Van Vliet-Vroegindeweij C, Van De Kamer JB, et al. FDG-PET and diffusion-weighted MRI in head-and-neck cancer patients: Implications for dose painting. Radiother Oncol 2013;106:250–4. 10.1016/j.radonc.2013.01.003. [DOI] [PubMed] [Google Scholar]
- [11].Ligtenberg H, Schakel T, Dankbaar JW, Ruiter LN, Peltenburg B, Willems SM, et al. Target volume delineation using diffusion-weighted imaging for MR-guided radiotherapy: a case series of laryngeal cancer validated by pathology. Cureus 2018:D. 10.7759/cureus.2465. [DOI] [PMC free article] [PubMed]
- [12].Schakel T, Peltenburg B, Dankbaar JW, Cardenas CE, Aristophanous M, Terhaard CHJ, et al. Evaluation of diffusion weighted imaging for tumor delineation in head-and-neck radiotherapy by comparison with automatically segmented 18F-fluorodeoxyglucose positron emission tomography. Phys Imaging Radiat Oncol 2018;5:13–8. 10.1016/j.phro.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Thorwarth D, Ege M, Nachbar M, Monnich D, Gani C, Zips D, et al. Quantitative magnetic resonance imaging on hybrid magnetic resonance linear accelerators : perspective on technical and clinical validation. Phys Imaging Radiat Oncol 2020;16:69–73. 10.1016/j.phro.2020.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kooreman ES, van Houdt PJ, Nowee ME, van Pelt VWJ, Tijssen RHN, Paulson ES, et al. Feasibility and accuracy of quantitative imaging on a 1.5 T MR-linear accelerator. Radiother Oncol 2019;133:156–62. 10.1016/j.radonc.2019.01.011. [DOI] [PubMed] [Google Scholar]
- [15].Kooreman ES, van Houdt PJ, Keesman R, van Pelt VWJ, Nowee ME, Pos F, et al. Daily Intravoxel Incoherent Motion (IVIM) in prostate cancer patients during MR-guided radiotherapy—a multicenter study. Front Oncol 2021;11:1–9. 10.3389/fonc.2021.705964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lawrence LSP, Chan RW, Chen H, Keller B, Stewart J, Ruschin M, et al. Accuracy and precision of apparent diffusion coefficient measurements on a 1.5 T MR-Linac in central nervous system tumour patients. Radiother Oncol 2021;164:155–62. 10.1016/j.radonc.2021.09.020. [DOI] [PubMed] [Google Scholar]
- [17].Yang Y, Cao M, Sheng K, Gao Y, Chen A, Kamrava M, et al. Longitudinal diffusion MRI for treatment response assessment: preliminary experience using an MRI-guided tri-cobalt 60 radiotherapy system. Med Phys 2016;43:1369–73. 10.1118/1.4942381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].van Houdt PJ, Yang Y, van der Heide UA. Quantitative magnetic resonance imaging for biological image-guided adaptive radiotherapy. Front Oncol 2021;10:1–9. 10.3389/fonc.2020.615643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].van Houdt PJ, Saeed H, Thorwarth D, Fuller CD, Hall WA, McDonald BA, et al. Integration of quantitative imaging biomarkers in clinical trials for MR-guided radiotherapy: conceptual guidance for multicentre studies from the MR-Linac Consortium Imaging Biomarker Working Group. Eur J Cancer 2021;153:64–71. 10.1016/j.ejca.2021.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kooreman ES, van Houdt PJ, Keesman R, Pos FJ, van Pelt VWJ, Nowee ME, et al. ADC measurements on the Unity MR-linac – a recommendation on behalf of the Elekta Unity MR-linac consortium. Radiother Oncol 2020;153:106–13. 10.1016/j.radonc.2020.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tijssen RHN, Philippens MEP, Paulson ES, Glitzner M, Chugh B, Wetscherek A, et al. MRI commissioning of 1.5T MR-linac systems – a multi-institutional study. Radiother Oncol 2019;132:114–20. 10.1016/j.radonc.2018.12.011. [DOI] [PubMed] [Google Scholar]
- [22].Zijlema SE, Tijssen RHN, Malkov VN, Van Dijk L, Hackett SL, Kok JGM, et al. Design and feasibility of a flexible, on-body, high impedance coil receive array for a 1.5 T MR-linac. Phys Med Biol 2019;64. 10.1088/1361-6560/ab37a8. [DOI] [PubMed]
- [23].Verhappen MH, Pouwels PJW, Ljumanovic R, Van Der Putten L, Knol DL, De Bree R, et al. Diffusion-weighted MR imaging in head and neck cancer: comparison between half-Fourier acquired single-shot turbo spin-echo and EPI techniques. Am J Neuroradiol 2012;33:1239–46. 10.3174/ajnr.A2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gao Y, Han F, Zhou Z, Cao M, Kaprealian T, Kamrava M, et al. Distortion-free diffusion MRI using an MRI-guided Tri-Cobalt 60 radiotherapy system: Sequence verification and preliminary clinical experience. Med Phys 2017;44:5357–66. 10.1002/mp.12465. [DOI] [PubMed] [Google Scholar]
- [25].Schakel T, Hoogduin JM, Terhaard CHJ, Philippens MEP. Technical Note: diffusion-weighted MRI with minimal distortion in head-and-neck radiotherapy using a turbo spin echo acquisition method: diffusion-weighted. Med Phys 2017;44:4188–93. 10.1002/mp.12363. [DOI] [PubMed] [Google Scholar]
- [26].Schick F SPLICE: Sub-second diffusion-sensitive MR imaging using a modified fast spin-echo acquisition mode. Magn Reson Med 1997;38:638–44. 10.1002/mrm.1910380418. [DOI] [PubMed] [Google Scholar]
- [27].Li Z, Pipe JG, Lee CY, Debbins JP, Karis JP, Huo D. X-PROP: a fast and robust diffusion-weighted propeller technique. Magn Reson Med 2011;66:341–7. 10.1002/mrm.23033. [DOI] [PubMed] [Google Scholar]
- [28].Srinivasan G, Rangwala N, Zhou XJ. Steer-PROP: a GRASE-PROPELLER sequence with interecho steering gradient pulses. Magn Reson Med 2018;79:2533–41. 10.1002/mrm.26898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Porter DA, Heidemann RM. High resolution diffusion-weighted imaging using readout-segmented echo-planar imaging, parallel imaging and a two-dimensional navigator-based reacquisition. Magn Reson Med 2009;62:468–75. 10.1002/mrm.22024. [DOI] [PubMed] [Google Scholar]
- [30].Miller KL, Pauly JM. Nonlinear phase correction for navigated diffusion imaging. Magn Reson Med 2003;50:343–53. 10.1002/mrm.10531. [DOI] [PubMed] [Google Scholar]
- [31].Verkooijen HM, Kerkmeijer LGW, Fuller CD, Huddart R, Faivre-Finn C, Verheij M, et al. R-IDEAL: a framework for systematic clinical evaluation of technical innovations in radiation oncology. Front Oncol 2017;7:1–7. 10.3389/fonc.2017.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].van Otterloo SRdM, Christodouleas JP, Blezer ELA, Akhiat H, Brown K, Choudhury A, et al. The MOMENTUM study: an international registry for the evidence-based introduction of MR-guided adaptive therapy. Front Oncol 2020;10:1328. 10.3389/fonc.2020.01328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hoogcarspel SJ, Zijlema SE, Tijssen RHN, Kerkmeijer LGW, Jürgenliemk-Schulz IM, Lagendijk JJW, et al. Characterization of the first RF coil dedicated to 1.5 T MR guided radiotherapy. Phys Med Biol 2018:63. 10.1088/1361-6560/aaa303. [DOI] [PubMed]
- [34].Shukla-Dave A, Obuchowski NA, Chenevert TL, Jambawalikar S, Schwartz LH, Malyarenko D, et al. Quantitative imaging biomarkers alliance (QIBA) recommendations for improved precision of DWI and DCE-MRI derived biomarkers in multicenter oncology trials. J Magn Reson Imaging 2019;49: e101–21. 10.1002/jmri.26518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].RSNA QIBA Diffusion-Weighted Imaging Task Force. QIBA Profile: Diffusion-Weighted Magnetic Resonance Imaging (DWI) 2019.
- [36].Barnhart HX, Barboriak DP. Applications of the repeatability of quantitative imaging biomarkers: a review of statistical analysis of repeat data sets. Transl Oncol 2009;2:231–5. 10.1593/tlo.09268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Paudyal R, Konar AS, Obuchowski NA, Hatzoglou V, Chenevert TL, Malyarenko DI, et al. Repeatability of quantitative diffusion-weighted imaging metrics in phantoms, head-and-neck and thyroid cancers: preliminary findings. Tomography 2019;5:15–25. 10.18383/j.tom.2018.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Habrich J, Boeke S, Nachbar M, Nikolaou K, Schick F, Gani C, et al. Repeatability of diffusion-weighted magnetic resonance imaging in head and neck cancer at a 1.5 T MR-Linac. Radiother Oncol 2022;174:141–8. 10.1016/j.radonc.2022.07.020. [DOI] [PubMed] [Google Scholar]
- [39].Boss MA, Chenevert TL, Waterton JC, Morris DM, Ragheb H, Jackson A, et al. Temperature-controlled isotropic diffusion phantom with wide range of apparent diffusion coefficients for multicenter assessment of scanner repeatability and reproducibility. Proc Int Soc Magn Reson Med 2014;22:4505. [Google Scholar]
- [40].Matoba M, Tuji H, Shimode Y, Toyoda I, Kuginuki Y, Miwa K, et al. Fractional change in apparent diffusion coefficient as an imaging biomarker for predicting treatment response in head and neck cancer treated with chemoradiotherapy. Am J Neuroradiol 2014;35:379–85. 10.3174/ajnr.A3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Khattab HM, Montasser MM, Eid M, Kandil A, Desouky SED. Diffusion-weighted magnetic resonance imaging (DWMRI) of head and neck squamous cell carcinoma: could it be an imaging biomarker for prediction of response to chemoradiation therapy. Egypt J Radiol Nucl Med 2020:51. 10.1186/s43055-020-00323-x. [DOI]
- [42].Siemann DW. The unique characteristics of tumor vasculature and preclinical evidence for its selective disruption by Tumor-Vascular Disrupting Agents. Cancer Treat Rev 2011;37:63–74. 10.1016/j.ctrv.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Forster J, Harriss-Phillips W, Douglass M, Bezak E. A review of the development of tumor vasculature and its effects on the tumor microenvironment. Hypoxia 2017;5:21–32. 10.2147/hp.s133231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Chavhan GB, Babyn PS, Thomas B, Shroff MM, Mark HE. Principles, techniques, and applications of T2*-based MR imaging and its special applications. Radiographics 2009;29:1433–49. 10.1148/rg.295095034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Weigel M, Helms G, Hennig J. Investigation and modeling of magnetization transfer effects in two-dimensional multislice turbo spin echo sequences with low constant or variable flip angles at 3 T. Magn Reson Med 2010;63:230–4. 10.1002/mrm.22145. [DOI] [PubMed] [Google Scholar]
- [46].Kerkmeijer LGW, Fuller CD, Verkooijen HM, Verheij M, Choudhury A, Harrington KJ, et al. The MRI-linear accelerator consortium: evidence-based clinical introduction of an innovation in radiation oncology connecting researchers, methodology, data collection, quality assurance, and technical development. Front Oncol 2016;6:215. 10.3389/fonc.2016.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].McDonald BA Chapter 5: DWI Pulse Sequence Optimization for Head and Neck on the MR-Linac. In “Development of Advanced MR-Guided Adaptive Radiation Therapy Methods for Head & Neck Cancers on the 1.5T MR-Linac.” The University of Texas MD Anderson Cancer Center Graduate School of Biomedical Sciences 2022. (99–121). 10.6084/m9.figshare.19521952.v2. [DOI]
- [48].Del Grande F, Santini F, Herzka DA, Aro MR, Dean CW, Gold GE, et al. Fat-suppression techniques for 3-T MR imaging of the musculoskeletal system. Radiographics 2014;34:217–33. 10.1148/rg.341135130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Obuchowski NA, Bullen J. Quantitative imaging biomarkers: effect of sample size and bias on confidence interval coverage. Stat Methods Med Res 2018;27:3139–50. 10.1177/0962280217693662. [DOI] [PubMed] [Google Scholar]
- [50].le Bihan D, Breton E, Lallemand D, Aubin M-L, Vignaud J, Laval-Jeantet M. Separation of diffusion and perfusion in intravoxel incoherent motion MR imaging. Radiology 1988;168:497–505. 10.1148/radiology.168.2.3393671. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


