Figure 1. Identification of a functional variant at the ZFHX3 locus on chromosome 16q22.
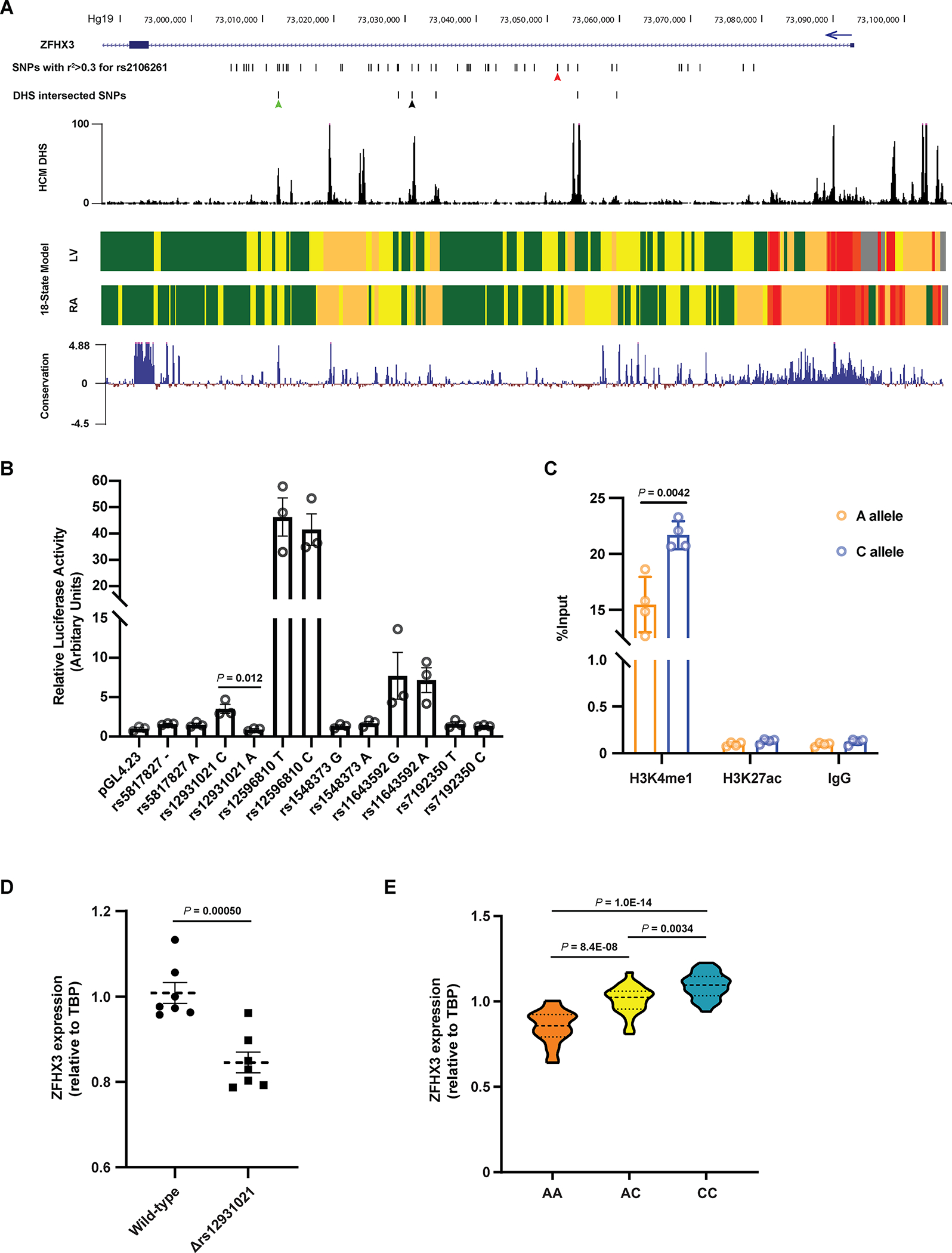
A. The region encompassing all common AF-associated SNPs with r2 > 0.3 with respect to the sentinel SNP rs2106261. The six candidate SNPs intersecting DNaseI hypersensitivity (DHS) signal are shown. The DHS signal in human cardiac myocytes and the mammalian conservation (Phylop) around this region were obtained from ENCODE. The 18-state models from Roadmap Epigenomics marking various regulatory elements in human left ventricles (LV) and right atria (RA) show regions of transcription, weak enhancer, active enhancer, transcription start site and repressed polycomb as green, yellow, orange, red and grey blocks, respectively. The red, green and black arrows indicate the position of rs2106261, rs12596810 and rs12931021, respectively. B. Luciferase data showing allele-specific activities for the six candidate SNPs in PSC-CMs. n = 3. C. Allele-specific ChIP-qPCR results in PSC-CMs heterozygous at rs12931021. The pulldown of H3K4me1 and H3K27ac were conducted to evaluate the enrichment of chromatin fragments containing either allele of rs12931021. n = 3. D. qPCR showing that deletion of rs12931021-containing regulatory element reduces the expression of ZFHX3 in PSC-CMs. n = 7. E. Relative expression of ZFHX3 in isogenic PSC-CMs carrying AA, AC or CC genotype at rs12931021. n = 24. P values are indicated (B-E). Data are mean ± s.e.m. Groups were compared using unpaired t tests (B-D). Groups were compared using ordinary one-way ANOVA with Tukey’s multiple comparisons test (E).
