Figure 2: Whole-Tissue Proteomics Finds Conservation of Extracellular Vesicle-Associated Processes Between Vascular and Valvular Disease Progression.
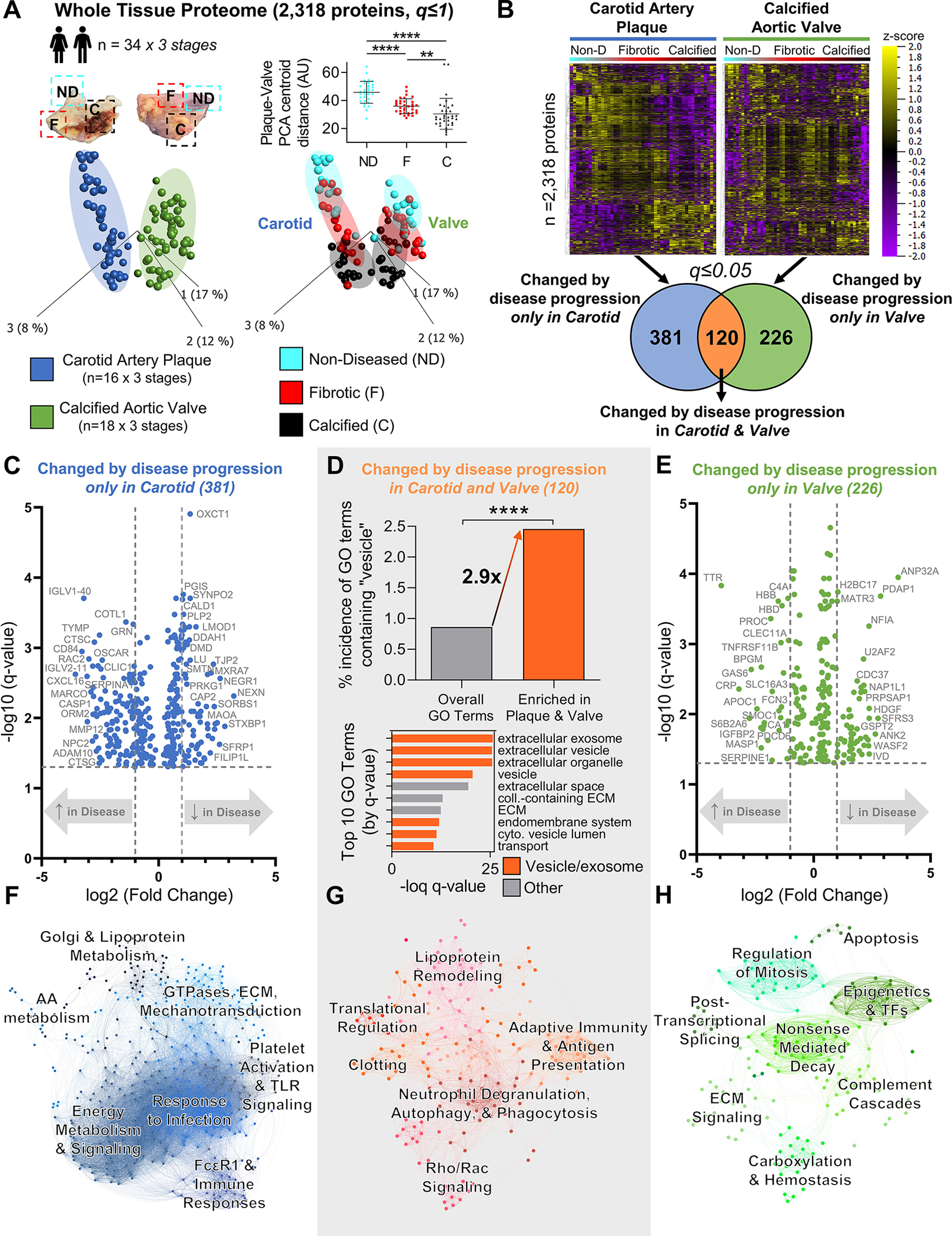
A, Principal component analysis of 2,318 proteins (q≤1) quantified by disease stage-specific whole-tissue proteomics of human carotid artery plaques and calcified aortic valves revealed significant convergence of carotid and valve tissues during disease pathogenesis; n=16 carotid artery plaque and 18 calcified aortic valve donors x 3 stages of disease (non-diseased, fibrotic, calcified) per donor; mean±SD; *p<0.05, ****p<0.0001. B, Unfiltered heat map analyses (q≤1; ordered by hierarchical clustering) demonstrated disease stage-specific alterations of protein abundances in carotid artery plaques (left) and calcified aortic valves (right). Enrichment analysis identified 381 proteins whose abundances were significantly differentially-enriched only by disease progression in carotid artery plaques, 226 proteins in calcified aortic valves, and 120 proteins in both tissue types (significantly-enriched proteins filtered at q≤0.05). C and E, Volcano plots of proteins significantly differentially-enriched by disease progression only in carotid artery plaques (381 proteins) or only in calcified aortic valves (226 proteins); cutoffs at a fold-change of 2 and a q-value of 0.05. D, In proteins significantly differentially-enriched in both carotid artery plaques and calcified aortic valves (120 proteins), we found a found a statistically significant 2.9-fold increase in the incidence of vesicle-associated GO terms vs. the total GO term database (****p<0.0001). In addition, 7 of the top 10 most-significant GO terms were associated with extracellular vesicles, exosomes, exocytosis, or secretion (orange bars). F-H, Networks based on KEGG, Reactome, and BioCarta pathway enrichment among proteins significantly differentially-enriched by disease progression only in carotid artery plaques (381 proteins, F), in both carotid artery plaques and calcified aortic valves (120 proteins, G), or only in calcified aortic valves (226 proteins, H) with pathways as the nodes (node size corresponds to -log(q-value)) and shared detected proteins between pathways as the edges (edge thickness matches the Jaccard index of overlap between detected proteins of the two connected pathway nodes). Unbiased clustering of pathways into real network communities by the Louvain method revealed shared- and tissue-specific drivers of disease pathogenesis.
