Figure 5: Sequencing Reveals the Human Tissue EV non-coding miRNAome from Vessels and Valves.
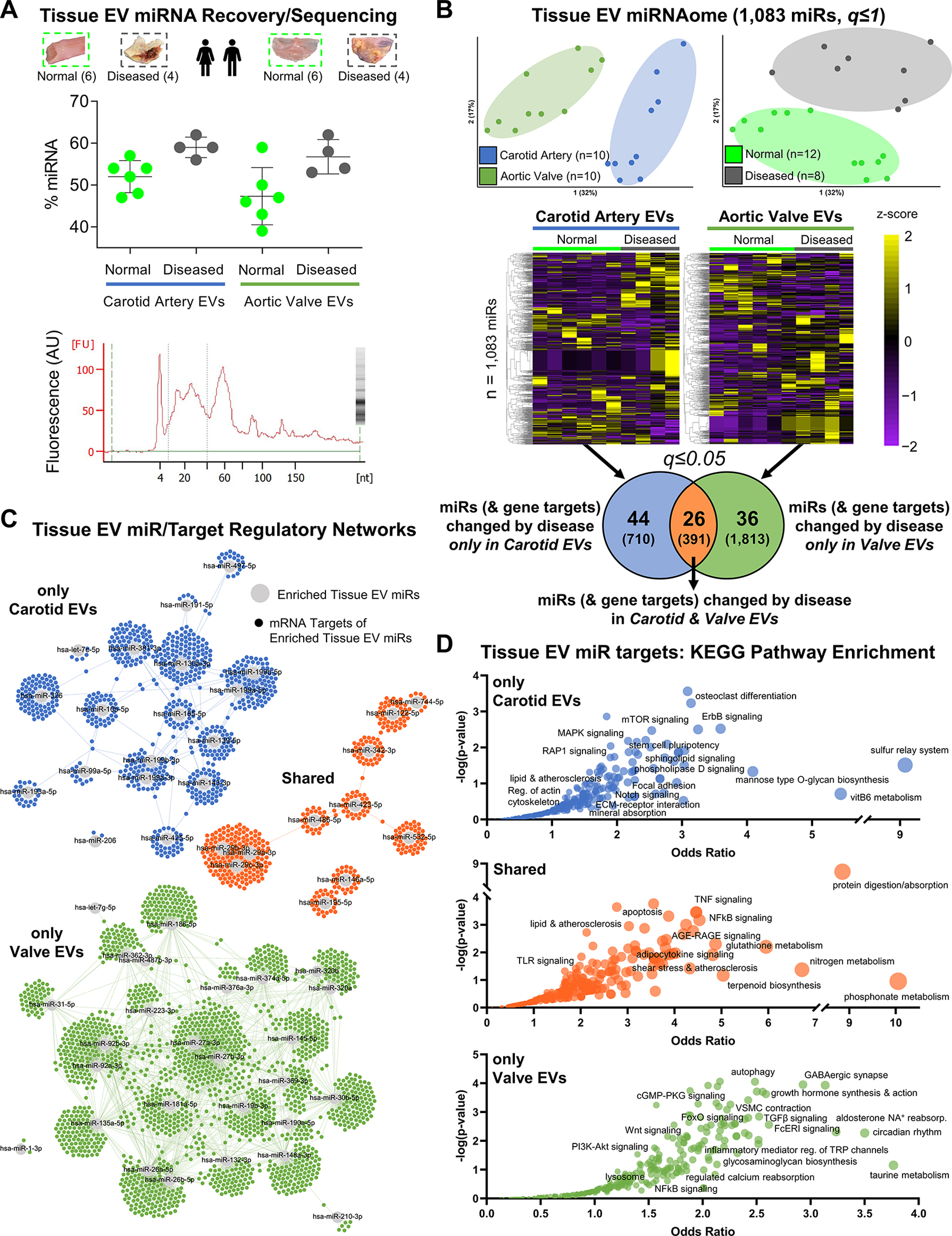
A, Along with proteomics, small RNA-seq was also performed on those tissue EVs collected from intact normal carotid arteries (n=6 donors), intact diseased carotid artery atherosclerotic plaques (n=4), intact normal aortic valves (n=6), and intact diseased calcified aortic valves (n=4). Top: RNA fragment analysis found that density gradient-enriched EVs contained enhanced levels of miRNA (mean miRNA content=53.0% of all small RNA; mean±SD). Bottom: representative fragment analysis tracing showing large miRNA-associated fragment peak at 24 nucleotides (nt). B, Unfiltered principal component analyses (q≤1) identified tissue- (left) and disease state-specific (right) clustering of 1,083 miRs sequenced in the tissue EV miRNAome. Unfiltered heat map analyses (q≤1; ordered by hierarchical clustering) characterized tissue EV miR cargoes altered between normal and diseased carotid arteries and aortic valves: 44 tissue EV miRs (with 710 unique high-confidence gene targets) were significantly differentially-enriched by disease pathogenesis only in carotid arteries, 36 tissue EV miRs (1,813 unique targets) changed only in aortic valves, and 26 tissue EV miRs (391 unique targets) were altered in both tissues (significantly-enriched miRs filtered at q≤0.05). C, miR/target regulatory networks of TargetScan-predicted gene targets (≥95th percentile weighted context++ score) of tissue EV miRs altered by disease only in carotid arteries (top; blue), in both carotid arteries and aortic valves (middle; orange), or only in aortic valves (bottom; green). D, Bubble plots of KEGG pathways significantly-enriched in the unique gene targets of tissue EV miRs changed by disease only in carotid arteries (top), in both carotid arteries and aortic valves (middle) or only in aortic valves (bottom) reveals the regulatory landscape associated with disease-altered tissue EV miR cargoes. Bubble size corresponds to the percentage of unique targets of differentially-enriched miRs amongst all pathway constituents.
