Figure 6: Pathway Networks Shared Across Omics Layers Identify Conserved Roles of Cardiovascular Tissue EV Cargoes Altered by Disease.
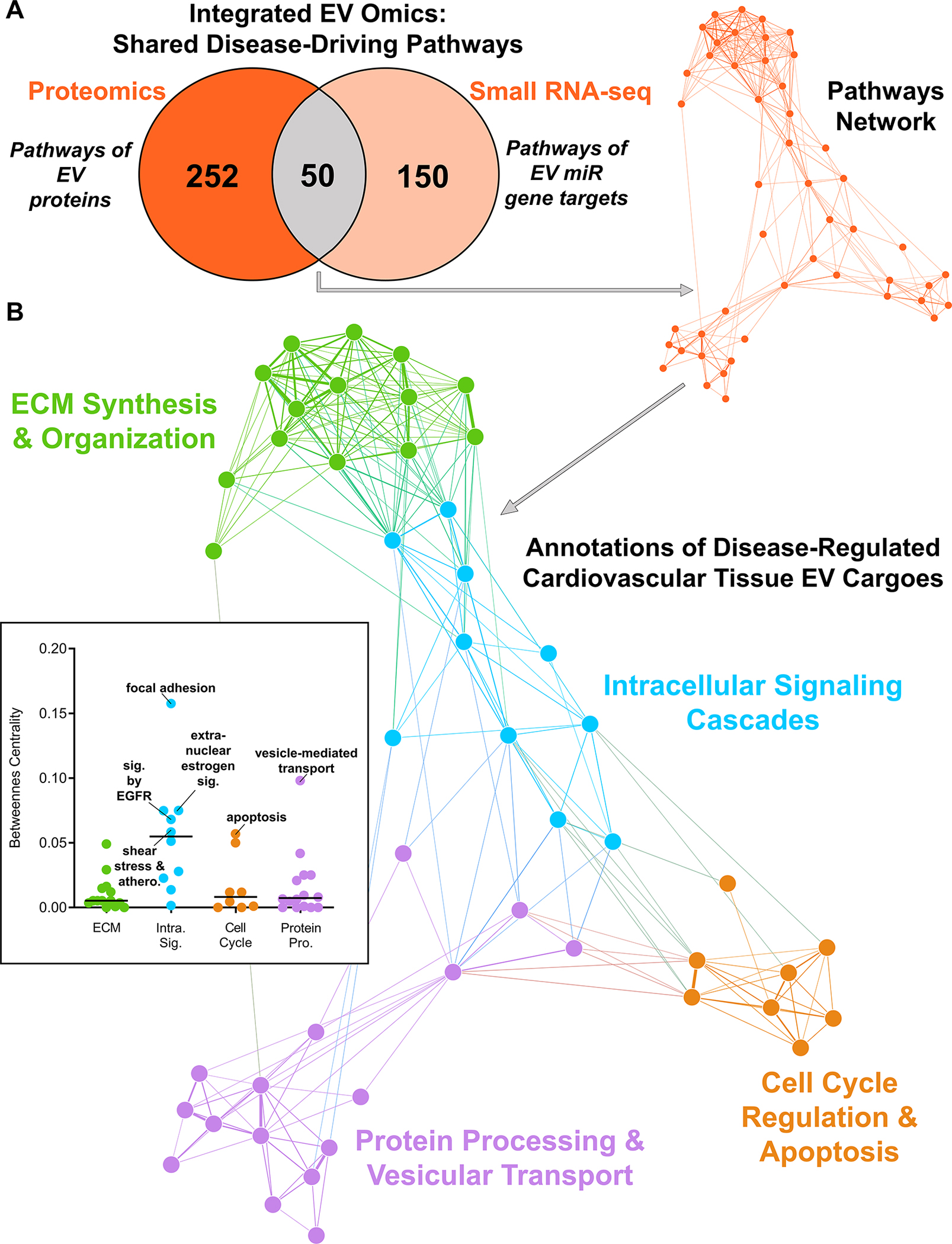
A, The network of 50 overlapping KEGG, Reactome, and BioCarta pathways that were significantly-enriched in the proteome and gene targets of miRs altered by disease in both intact carotid artery and aortic valve tissue EVs (n=6 normal carotid arteries, n=4 diseased carotid artery atherosclerotic plaques, n=6 normal aortic valves, n=4 diseased calcified aortic valves). Pathways are nodes (node size corresponds to -log(q-value)) and shared detected genes between pathways are edges (edge thickness matches the Jaccard index of overlap between detected genes of the two connected pathway nodes). B, Louvain clustering revealed 4 distinct annotations shared by disease-altered cardiovascular tissue-derived EV cargoes, including modulation of intracellular signaling cascades and cell cycle regulation and apoptosis. Pathway network betweenness-centrality scores (inset) implicate estrogen/epidermal growth factor signaling as a key common molecular constituent of tissue EVs.
