Fig. 5: Loss of FH activity disrupts de novo purine biosynthesis.
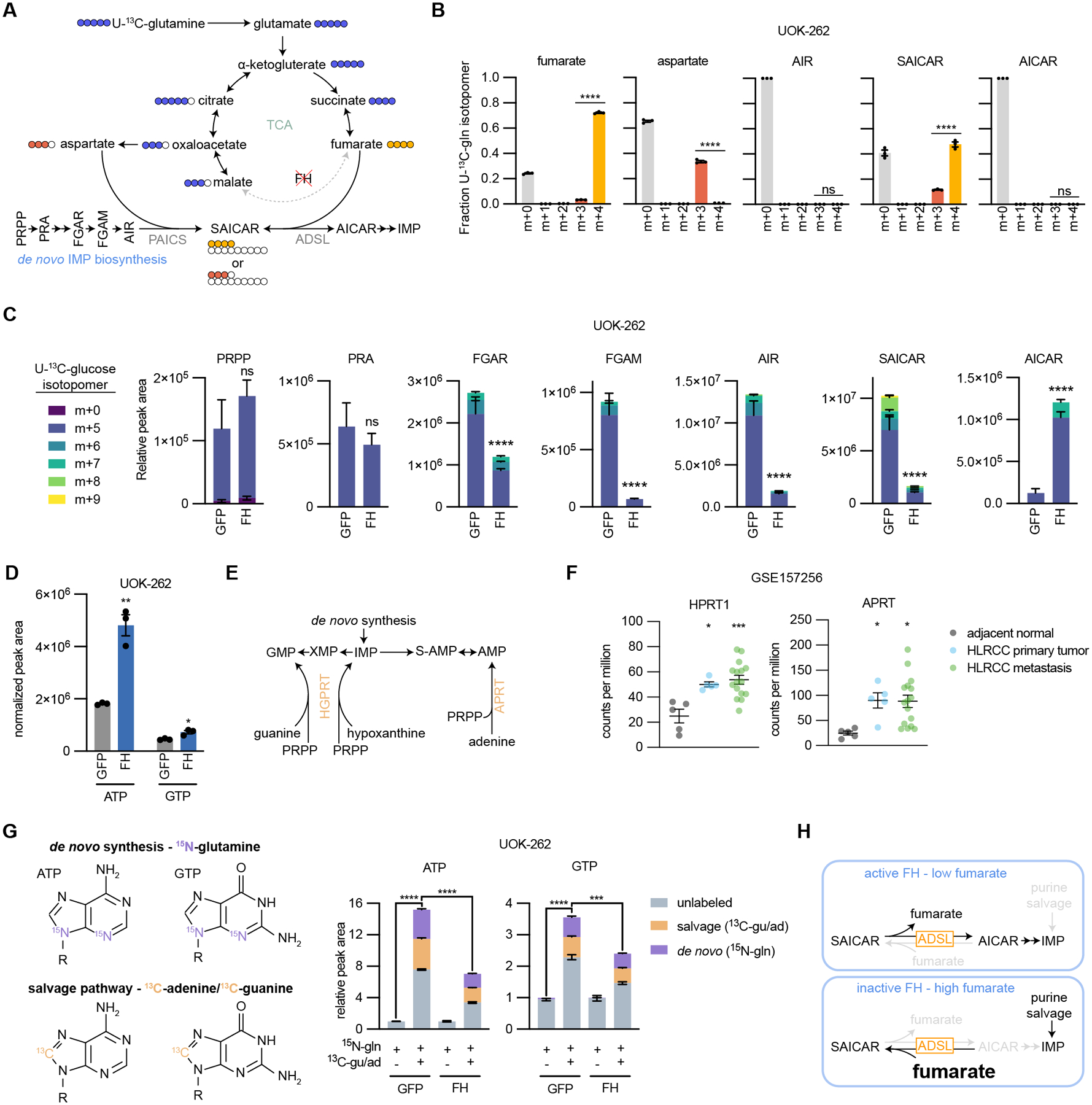
(A) Schematic representing the flow of carbons in the TCA cycle and de novo IMP biosynthesis from U-13C-glutamine in HLRCC cells. Fraction of 13C-labeled isotopomers for (B) fumarate, aspartate, AIR, SAICAR, and AICAR. (C) Relative peak areas for de novo IMP biosynthesis intermediates in UOK-262 cells treated with U-13C-glucose. Bars are colored by the relative amount of individual 13C isotopomers. (D) Normalized peak areas for ATP and GTP. (E) Schematic of the purine salvage pathway. (F) Expression of APRT and HPRT1 in HLRCC patient primary tumors, metastases, and adjacent normal kidney tissue. Data is publicly available at NCBI Gene Expression Omnibus GSE157256. (G) Depiction of how ATP and GTP are labeled by amide-15N-glutamine, 8-13C-adenine, and 8-13C-guanine. UOK-262 cells were treated for 6 hours with 4 mM amide-15N-glutamine, 50 μM 8-13C-adenine, and 50 μM 8-13C-guanine, and ATP and GTP were analyzed by LC-MS for mass shifts corresponding to de novo synthesis (purple) or salvage pathway (orange). In order to determine the capacity for salvage pathway activity, each group was normalized to 15N-glutamine only treated samples. (H) Summary of how FH loss impacts purine biosynthesis. In cells without active FH, fumarate accumulates which drives the succination of AICAR by ADSL to produce SAICAR (reverse reaction). This prevents de novo IMP biosynthesis, leaving the cells reliant on the salvage pathway to support purine synthesis.
