Figure 3. Bromodomain dispensability conserved across cell lines.
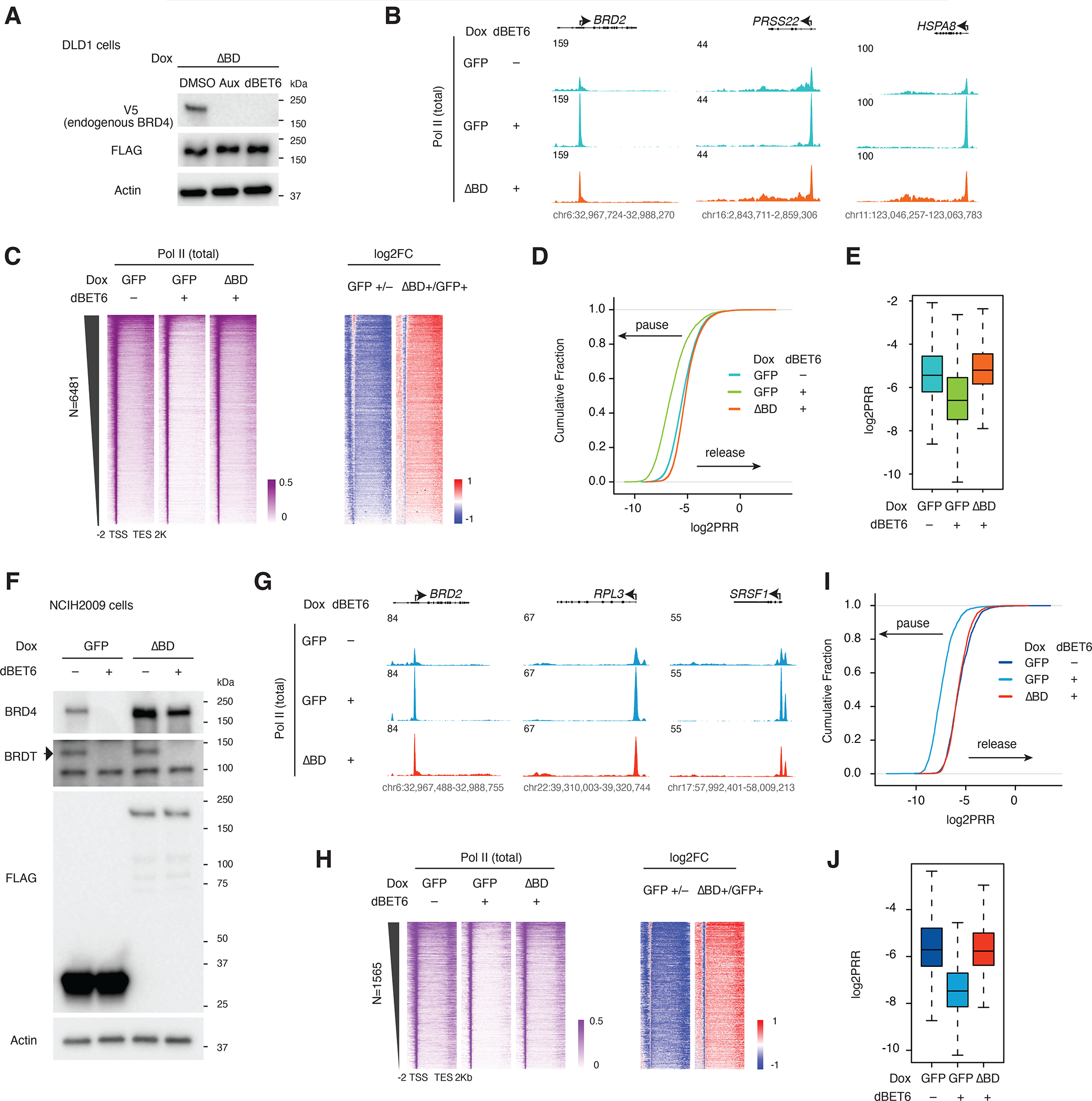
A. Western blot showing degradation of endogenous BRD4 (V5) but not the bromodomain-less BRD4 mutant (FLAG) upon auxin or dBET6 treatment in the BRD4-IAA7 DLD1 degron line.
B. Track examples of the total Pol II ChIP-seq signal at the BRD2, PRSS22 and HSPA8 loci after dBET6 treatment and rescue by bromodomain-less BRD4 in DLD-1 cells.
C. Heatmap showing the genome-wide Pol II occupancy and fold change in occupancy for the bromodomain-less mutant construct vs GFP control after dBET6 treatment in DLD-1.
D. ECDF of the log2PRR from the total Pol II ChIP-seq for the dBET6 rescue experiment in DLD-1.
E. Boxplot of the log2PRR from the total Pol II ChIP-seq for the dBET6 rescue experiment in DLD-1.
F. Western blot showing the depletion of endogenous BRD4 and BRDT, but not the bromodomain-less mutant, by dBET6 treatment (3h) in NCIH2009 cells.
G. Track examples of the total Pol II ChIP-seq signal upon dBET6 treatment (3h) and rescue by bromodomain-less BRD4 in NCIH2009 cells.
H. Heatmap showing the Pol II occupancy profiles and corresponding fold changes across the 1565 genes for which dBET6 causes strong pausing (2-fold reduction of Pol II at gene bodies and 2-fold reduction of PRR), rescued by bromodomain-less BRD4 in NCIH2009.
I. ECDF of the log2PRR from the Pol II ChIP-seq for the dBET6 rescue experiment in NCIH2009 (N=1565).
J. Boxplot of the log2PRR from the Pol II ChIP-seq for the dBET6 rescue experiment in NCIH2009 (N=1565).
