Figure 4. BRD4 C-terminal fragment interacts with PTEFb and rescues Pol II pause release.
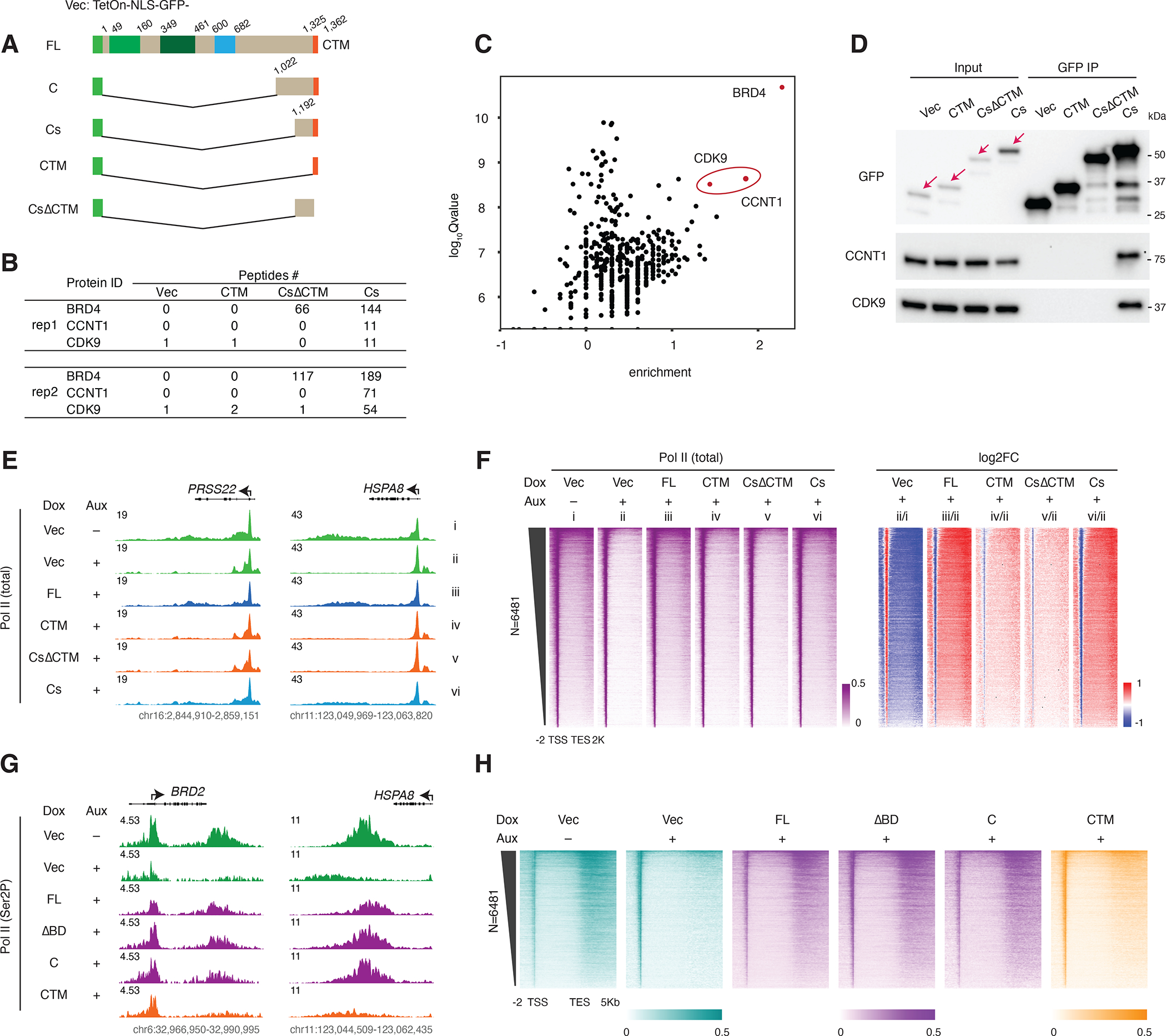
A. Schematic of the GFP-tagged FL BRD4 and BRD4 C-terminus constructs (vector: TetOn-NLS-GFP). Cs: short C terminus.
B. GFP IP-MS results showing peptide counts for the bait (BRD4) and PTEFb complex components (CCNT1 and CDK9) in two replicates after Dox (50nM) induced expression of Vector, CTM, CsΔCTM or Cs mutant constructs for 2 days. Note: the 0 bait peptide count for the CTM construct is probably due to impaired peptide alignment, as the CTM construct is itself a (GFP-tagged) peptide of only 37 residues.
C. Scatter plot showing the log10Qvalue vs the enrichment over Vector (calculated by log10FC of peptide counts +1) for the BRD4-Cs interacting proteins in the second GFP IP-MS replicate.
D. Western blot validating the GFP IP-MS result using the elute from the second replicate.
E. Track examples for total Pol II ChIP-seq signal at the PRSS22 and HSPA8 gene loci upon auxin treatment and rescue by different GFP-tagged mutants.
F. Heatmap showing the genome-wide Pol II occupancy profile and the corresponding fold changes for the GFP-tagged rescue experiment.
G. Track examples for Ser2P ChIP-seq signal at the BRD2 and HSPA8 gene loci upon auxin treatment and rescue by different GFP-tagged mutants.
H. Heatmap showing the genome-wide Pol II occupancy profile and the corresponding fold changes for the GFP-tagged rescue experiment.
