Figure 5. Identification of a distinct BRD4-PTEFb population: active but not histone acetylation-bound.
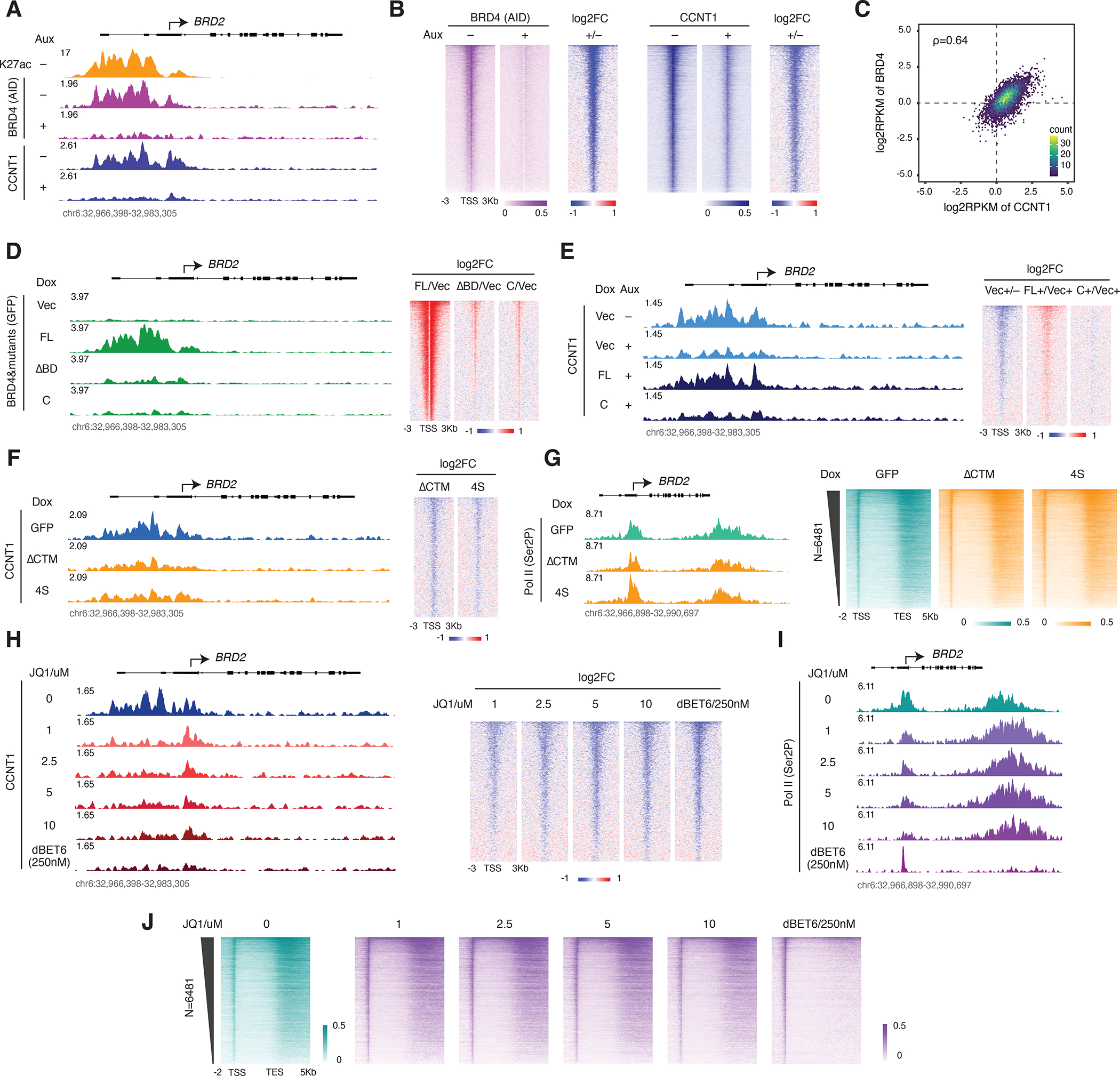
A. Track examples for AID ChIP-seq in the BRD4-AID line and CCNT1 ChIP-seq in the BRD4-IAA7 line with/without auxin treatment (3h). H3K27ac ChIP-seq signal in the BRD4-AID line is shown for comparison.
B. Heatmap showing the genome-wide BRD4 and CCNT1 occupancy profile and the corresponding fold change upon auxin treatment for the conditions in A. Genes ranked by BRD4 signal in control.
C. Scatter plot showing the relative promoter density of CCNT1 vs BRD4, calculated by the log2RPKM of the ChIP-seq signals at the regions flanking (±1Kb) the Pol II pausing sites.
D. Track examples for GFP ChIP-seq signal upon induction of GFP-tagged BRD4-FL or the indicated mutants (left). Heatmap showing the genome-wide fold changes of the GFP-tagged BRD4 mutants relative to the vector (right). The endogenous BRD4 was depleted by auxin treatment (3h) in all samples. Genes are ranked by GFP signal in the GFP-BRD4-FL condition.
E. Track examples for CCNT1 ChIP-seq signal upon endogenous BRD4 depletion and rescue with indicated GFP-tagged mutants (left). Heatmap showing the genome-wide CCNT1 fold changes (right). Genes are ranked by CCNT1 signal in the untreated vector control.
F. Track examples for the CCNT1 ChIP-seq signal upon induction of the FLAG-tagged, CTM-less BRD4 mutants by Dox treatment for 2 days (left). Heatmap showing the genome-wide profile of fold changes in CCNT1 occupancy (right). Genes are ranked by CCNT1 signal in the GFP control.
G. Track examples for the Ser2P ChIP-seq from the same samples in F (left). Heatmap showing the corresponding genome-wide Ser2P occupancy profile (right).
H. Track examples for the CCNT1 ChIP-seq in a series concentration of JQ1 and 250nM dBET6 treatment for 3h in the BRD4-IAA7 cells (left). Heatmap showing the corresponding genome-wide CCNT1 fold changes relative to DMSO treatment (right). Genes ranked by CCNT1 signal in the DMSO control.
I. Track examples for the Ser2P ChIP-seq from the same JQ1 or dBET6-treated samples in H.
J. Heatmap showing the corresponding genome-wide Ser2P occupancy profile.
