Figure 2. Cryo-EM structures of PG9RSH and PGT145 variants in complex with BG505 DS-SOSIP Env trimer.
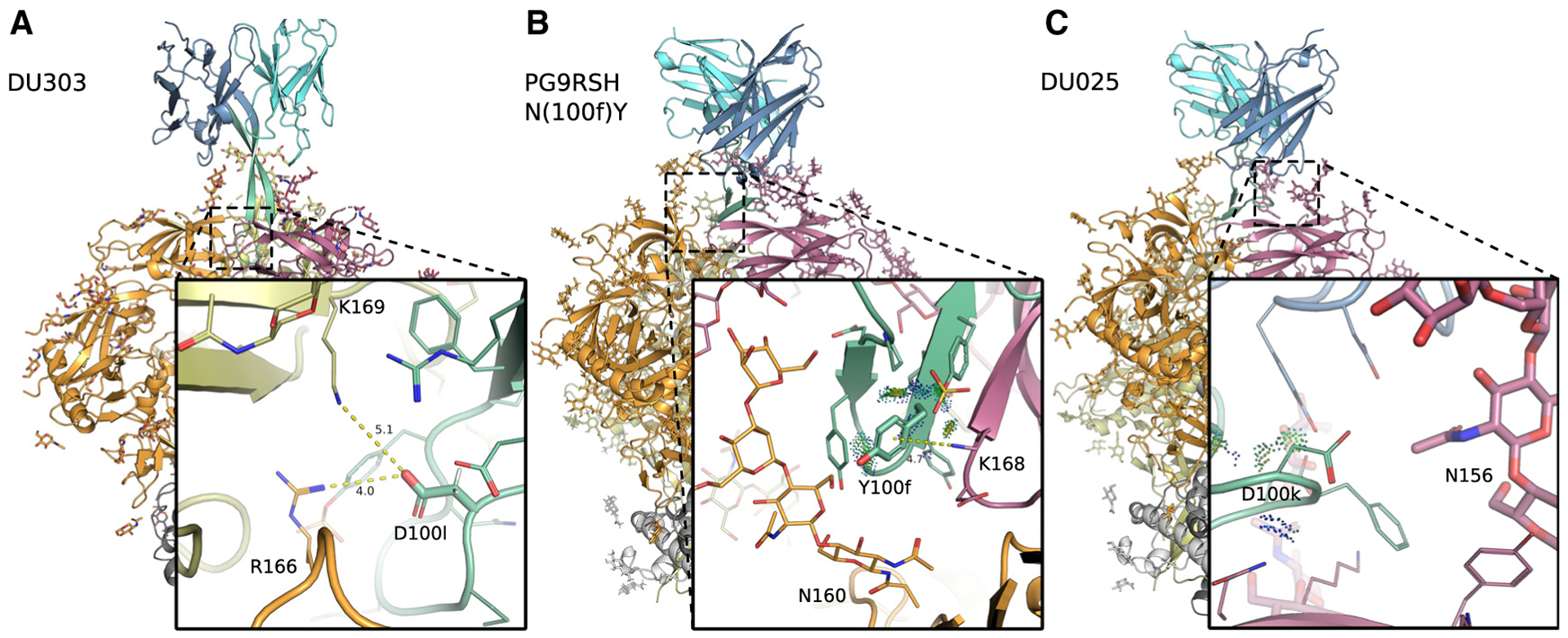
Backbone shown in ribbon representation with glycans, and amino acids shown as sticks or lines. Env subunits colored with warm colors and grays, and antibodies shown in cool colors. CDRH3 (residues 95–102) is shown in green. Distances (Å) are shown with dotted yellow lines, and energetic interactions are shown with Probe dots. Members of the PG9RSH (A and B) and PGT145 lineages (C) interact with the trimer apex and are characterized by a negatively charged CDRH3 that is hammer-like or extended, respectively.
(A) PGT145 mutation N(100l)D forms more favorable electrostatic interactions with gp120.
(B) The PG9RSH N(100f)Y mutation creates interactions with a side-chain nitrogen from gp120 residue K168, forming geometry consistent with a π-cation interaction.
(C) The PG9RSH Y(100k)D mutation forms long-range interactions with polar and positively charged residues Q170 and K305.
