Figure 3. OSPREY design ensembles correctly predicted structural features for PG9RSH and PGT145 variants.
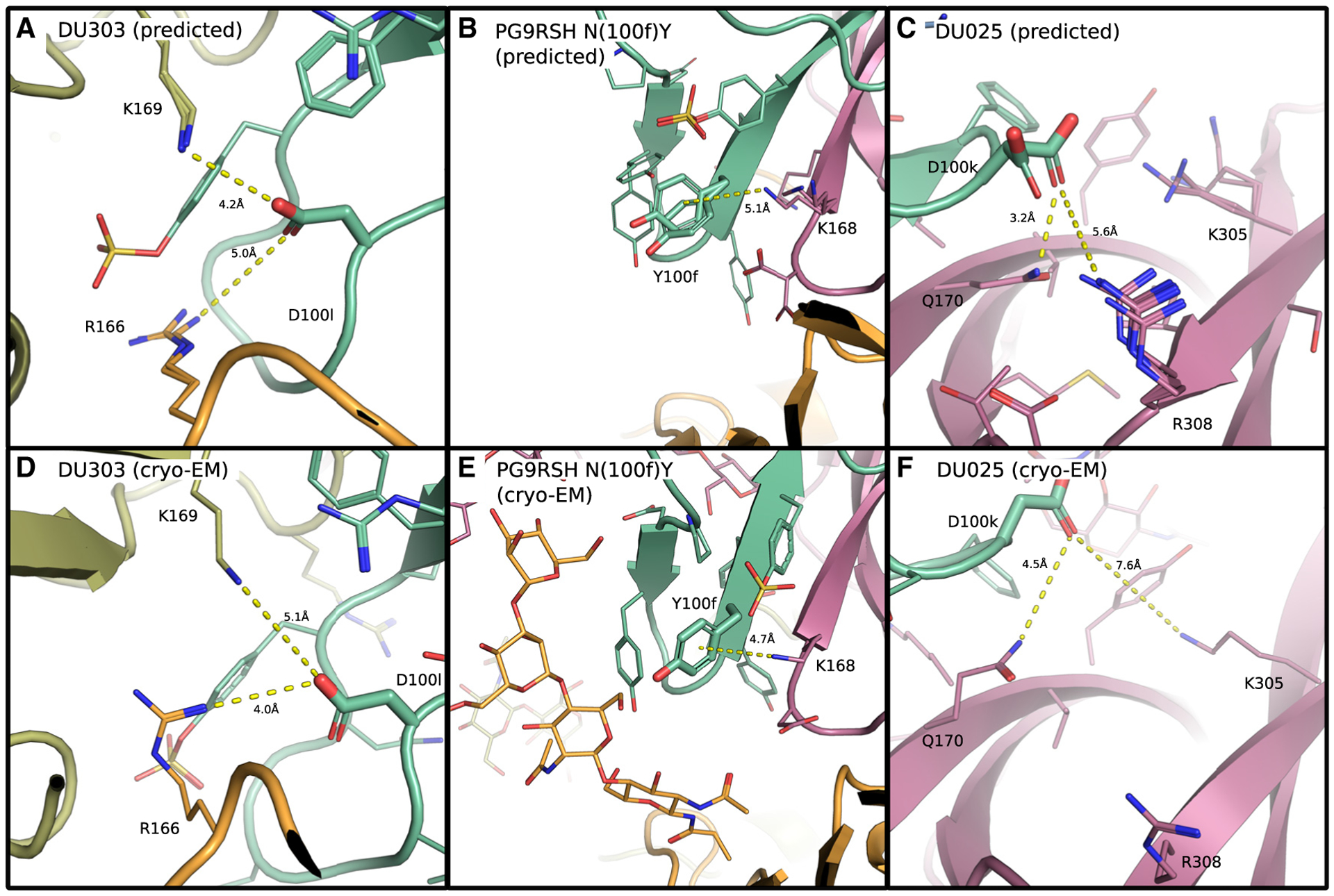
Ten members of the low-energy ensemble (LEE) predicted by OSPREY are shown for variants of PGT145 (A) and PG9RSH (B and C) above corresponding cryo-EM structures (D–F). Backbones are shown as ribbons with amino acids shown as lines or as sticks with Env subunits colored with warm colors, and antibody CDRH3 loops (residues 95–102) are shown in green. Distances (Å) are shown with dotted yellow lines.
(A) PGT145 mutation N(100l)D is predicted to form electrostatic interactions with gp120 residues R166 and K169. A carboxyl oxygen of D(100l) lies 5 and 4.2 Å from side-chain nitrogens of gp120 residues R166 and K169, respectively. Despite a lateral translation of the CDRH3 loop relative to the trimer apex, the LEE correctly predicts features of the experimental structure (F).
(B) The PG9RSH N(100f)Y mutation creates interactions with gp120 residue K168. The side-chain amino nitrogen of K168 lies 5.1 Å from the ring plane of Y(100f), forming geometry consistent with a π-cation interaction. The LEE correctly predicts interactions found in the experimentally determined structure (D).
(C) The PG9RSH Y(100k)D mutation is predicted to form electrostatic interactions with polar and charged residues on gp120. A carboxyl oxygen of D(100k) lies 3.2 Å from the side-chain nitrogen of Q170 and 5.6 Å from R308. The LEE correctly predicts interactions with Q170, but a translation and rotation of the CDRH3 loop places R308 further away (E).
