Fig. 4. OBOX binding in 2C embryos.
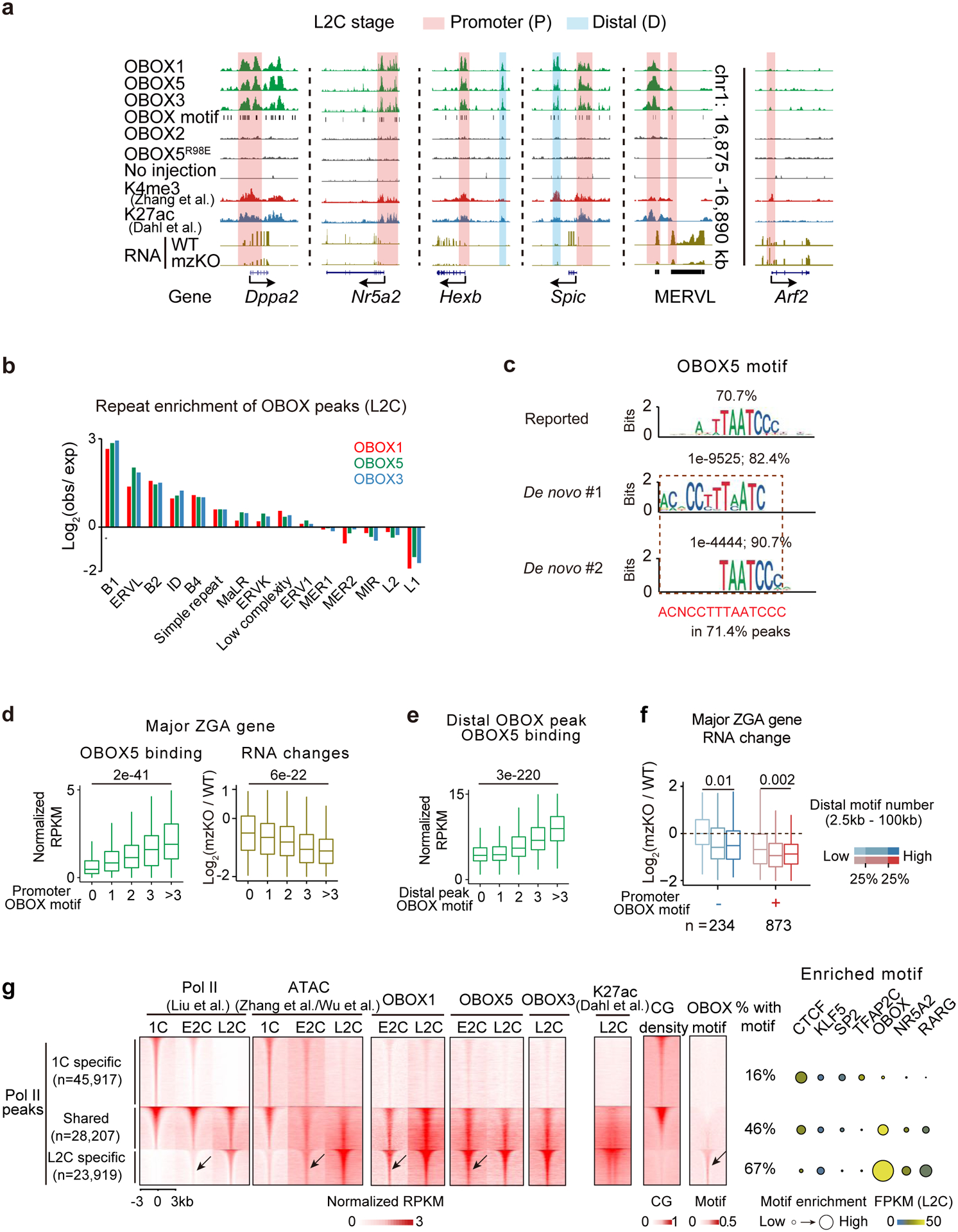
a, The UCSC browser snapshots showing OBOX1/5/3 binding at example genes and repeats in L2C embryos. Stacc-seq of OBOX2, OBOX5R98E binding, and Stacc-seq in embryos without injection are negative controls. H3K4me328, H3K27ac42, OBOX motif, and RNA levels in WT and Obox mzKO embryos are also shown. b, Bar chart showing repeat enrichment at OBOX binding peaks at L2C. c, Reported motif27, de novo top 1, 2, and combined extended motif identified by OBOX5 binding peaks in embryos. The percentages of peaks containing these motifs and P-values are shown. d, Box plots showing OBOX5 binding at major ZGA gene promoters in WT L2C embryos (left) and the major ZGA gene expression fold-changes upon OBOX depletion (right, 3 biological replicates). P-value, two-sided Wilcoxon rank-sum test. 234, 272, 232, 169, and 201 genes have 0, 1, 2, 3, and >3 OBOX motifs on promoters, respectively. Centre line, median; box, 25th and 75th percentiles; whiskers, 1.5 × IQR (same for d and e). e, Box plot showing OBOX5 binding enrichment at distal binding peaks in L2C embryos. P-value, two-sided Wilcoxon rank-sum test. 5,885, 16,226, 5,314, 1,370, and 561 distal OBOX3 binding peaks have 0, 1, 2, 3, and >3 OBOX motifs, respectively. f, Box plots showing ZGA gene expression changes upon Obox knockout (3 biological replicates). P-value, two-sided Wilcoxon rank-sum test. n, major ZGA gene number. g, Left, heatmaps showing Pol II binding9, chromatin accessibility (ATAC)26,31, OBOX binding, H3K27ac42, CG density and OBOX motif enrichment at 1C-specific, shared, and L2C-specific Pol II peaks in WT embryo. The percentages of peaks with at least one OBOX motif are shown. The arrows indicate Pol II, accessible chromatin, OBOX1/5 binding, and OBOX motif at the L2C-specific Pol II peaks in the E2C embryos. Right, enrichment of known transcription factor motifs. Motif P-value, area of the circle.
