SUMMARY
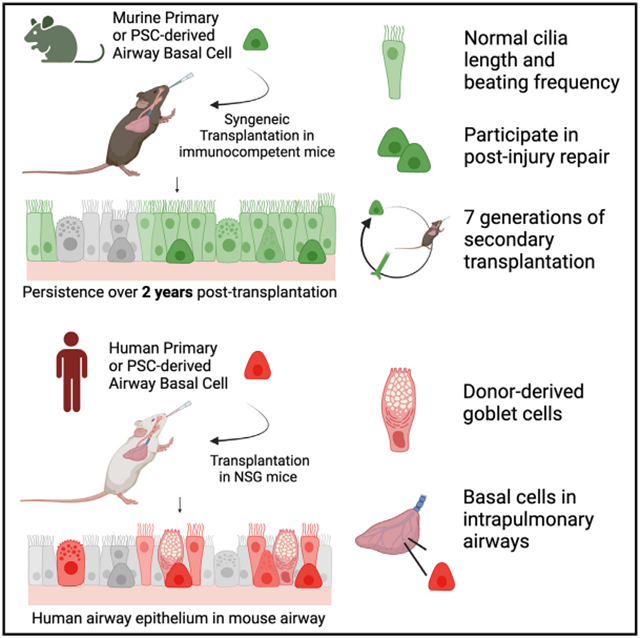
Life-long reconstitution of a tissue’s resident stem cell compartment with engrafted cells has the potential to durably replenish organ function. Here, we demonstrate the engraftment of the airway epithelial stem cell compartment via intra-airway transplantation of mouse or human primary and pluripotent stem cell (PSC)-derived airway basal cells (BCs). Murine primary or PSC-derived BCs transplanted into polidocanol-injured syngeneic recipients give rise for at least two years to progeny that stably display the morphologic, molecular, and functional phenotypes of airway epithelia. The engrafted basal-like cells retain extensive self-renewal potential, evident by the capacity to reconstitute the tracheal epithelium through seven generations of secondary transplantation. Using the same approach, human primary or PSC-derived BCs transplanted into NOD scid gamma (NSG) recipient mice similarly display multilineage airway epithelial differentiation in vivo. Our results may provide a step toward potential future syngeneic cell-based therapy for patients with diseases resulting from airway epithelial cell damage or dysfunction.
Graphical Abstract
In brief
Kotton and colleagues demonstrate long-term reconstitution of the mouse airway basal stem cell compartment in vivo via the engraftment of cultured primary or pluripotent stem cell-derived mouse or human airway basal cells. This may serve as a first step toward future cell-based therapies for airway epithelial diseases.
INTRODUCTION
Adult mammalian epithelial tissues are maintained throughout life through either the proliferation of common mature cells or the self-renewal and differentiation of tissue-resident stem cells.1-6 Engraftment of exogenous epithelial stem cells provides an opportunity to potentially supplement or replenish endogenous reparative cells and has been attempted with success for ectodermally derived epithelia.7-9 However, similar engraftment has been challenging to achieve for internal tissues with rare exceptions.10,11 This in turn has prevented successful long-lived rescue of diseased internal epithelia, such as the respiratory air-ways.12 Reconstituting the tissue-resident stem cell compartment of internal epithelial tissues has the theoretical advantage of resulting in durable and functional engraftment, since stem cells can undergo multipotent differentiation as well as self-renewal in vivo, if successfully engrafted.
The conducting airways of the lung present a particularly attractive target for cell transplantation, since: (1) the entire airway epithelium is in contact with the outside environment, providing potential access for exogenous cells; (2) the resident stem cells of the pseudostratified respiratory epithelium, basal cells (BCs), are known and have been extensively characterized6,13-20; and (3) genetic diseases that affect the airway, such as cystic fibrosis or primary ciliary dyskinesia, remain significant sources of morbidity and mortality, which can be potentially targeted by engraftment of gene-corrected airway stem cells. Yet, the BC compartment of the airways remains frustratingly difficult to target with cells or genes, as it is protected by both luminal cells that abut the airway surface and a highly effective innate immune system that have evolved to protect the airway epithelium from being penetrated by inhaled exogenous cells, pathogens, or particles.
While transplantation of exogenous cells, such as primary cells harvested from fetal or adult lungs, into the airway or alveolar epithelium of immunodeficient mouse recipients has been successful,21-28 durable reconstitution of the airway stem cell compartment has yet to be demonstrated. Primary airway BCs and their equivalents generated from pluripotent stem cells (PSCs) in vitro both represent compelling cell types for transplantation, since they can be maintained and expanded in cell culture to generate the large numbers of cells with retained stem cell phenotype that would be required for transplantation. Furthermore, both cell types can be stored as frozen archives or genetically manipulated,29features that would be expected to facilitate future cell-based therapies.
Here, we reconstitute the airway stem cell compartment of mice by engraftment of mouse or human primary or PSC-derived basal stem cells (primary BC or iBC). After airway epithelial injury with polidocanol, we observe that transplantation of murine BCs or iBCs results in engrafted BCs with self-renewal and multiline-age differentiation capacity, the defining features of airway basal stem cells. iBCs engrafted in recipient syngeneic mice can persist for more than two years and can be serially transplanted through seven generations of secondary mouse recipients. Furthermore, as a preclinical test, we demonstrate in vivo engraftment of injured NOD scid gamma (NSG) mouse recipient airway epithelium with human primary BCs and human iBCs. Collectively, our results establish primary and PSC-derived BC transplantation as a potential method for in vivo reconstitution of an internal epithelial tissue-resident stem cell compartment and may serve as a first step toward future autologous cell-replacement therapy for patients with airway diseases.
RESULTS
Generation and maintenance of murine primary and PSC-derived basal cells in vitro
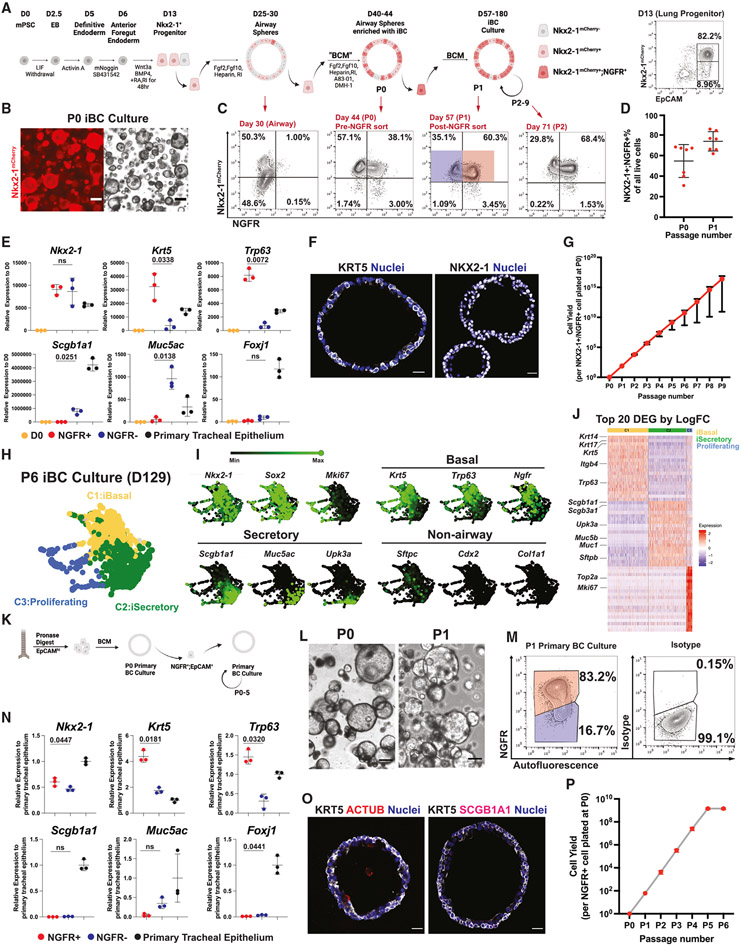
To develop an approach for reconstituting the resident stem cell compartment of the murine trachea, we first sought to develop a common serum-free, feeder-free medium that would allow both primary mouse BCs and iBCs to be expanded in cell cultures into the large cell numbers likely to be needed for transplantation. We adapted our previously published methods for derivation of a diversity of lung epithelial lineages, including human iBCs,29-36 and optimized conditions for similarly differentiating mouse PSCs into iBCs (Figure 1A). We first differentiated mycoplasma-free, karyotypically normal (Figure S1A) mouse PSCs carrying an Nkx2-1mCherry reporter37,38 into definitive endoderm, patterned the endoderm into anterior foregut,30 induced lung specification into Nkx2-1mCherry+ primordial progenitors,35,36 and generated proximalized airway epithelial cells in 28 days, as we previously published.32,36 We then purified Nkx2-1mCherry+ cells at this stage for three-dimensional (3D) culture in Matrigel in a newly optimized “basal cell medium” (hereafter “BCM”), composed of our published fibroblast growth factor (FGF)-supplemented airway media32,36 with added inhibitors of mothers against decapentaplegic homolog (SMAD)-dependent bone morphogenetic protein (BMP) and transforming growth factor beta (TGF-β) signaling, as suggested by prior reports on their effects on primary BCs.39,40 Exposure of PSC-derived airway organoids to BCM induced upregulation of the BC surface protein marker, nerve growth factor receptor (NGFR) (Figures 1B-1D). By day 40–44, Nkx2-1mCherry+ monolayered epithelial spheres emerged, with 54.73% ± 16.00% of cells co-expressing Nkx2-1mCherry and NGFR (Figures 1B-1D). After single-cell dissociation, passaging, and further expansion as epithelial spheres in BCM, the percentage of Nkx2-1mCherry+/NGFR+ cells increased to 73.86% ± 9.42% (Figure 1D).
Figure 1. Generation and expansion in culture of murine iBC and primary BC culture in defined serum-free medium.
(A) Schematic of in vitro directed differentiation of murine pluripotent stem cells (PSCs) carrying an Nkx2-1mCherry reporter into airway basal epithelial-like cells (iBC), followed by their expansion as monolayered epithelial spheres in serially passaged 3D serum-free, feeder-free Matrigel cultures. RA, retinoic acid; RI, ROCK inhibitor Y27632; BCM, basal cell medium. Representative day 13 fluorescence-activated cell sorting (FACS) plot is shown.
(B) Representative iBC culture (day 52) at passage 1 (P1) after sorting NGFR+/Nkx2-1mCherry+ cells. Scale bars, 200 μm.
(C) Representative FACS plots at different stages of the iBC-directed differentiation protocol shown in (A). Day 57 (P1 iBC) samples were analyzed by RT-qPCR in (E), with red sorting gate representing NGFR+ samples and blue sorting gate representing NGFR− samples.
(D) Quantification of percent of cells co-expressing Nkx2-1mCherry and NGFR+ cells out of all live cells analyzed by FACS at P0 and P1, respectively. Bars indicate mean ± SD. n = 7 biological replicates.
(E) Reverse-transcription quantitative PCR (RT-qPCR) analysis of P1 iBC culture. Statistical analysis performed by paired, two-tailed Student’s t test. Bars indicate mean ± SD. n = 3 biological replicates.
(F) Immunofluorescence confocal microscopy of paraffin-embedded tissue sections of representative iBC epithelial spheres stained for NKX2-1 and KRT5 proteins. Scale bars, 20 μm. Blue nuclei are counterstained with Hoechst33342.
(G) Quantification of iBC cell yield and growth kinetics over nine passages. Mean ± SD are shown for three replicated differentiations separated at day 0.
(H) Dimensionality reduction visualization by SPRING plots of iBC global transcriptomes profiled by scRNA-seq. Outgrowth of iBC cultures after six serial passages (P6; day 129) were analyzed by Louvain clustering (resolution = 0.5), and cell clusters were annotated based on canonical markers. iBasal, PSC-derived basal-like cell; iSecretory, PSC-derived secretory-like cell.
(I) Selected canonical marker gene expression levels for each indicated marker shown overlaid on the SPRING plot from (H).
(J) Heatmap of the top 20 differentially expressed genes (DEGs) ranked by log fold change (logFC) in each of the three cell clusters (C1–C3) annotated in (H): iBasal, iSecretory, and Proliferating cluster.
(K) Schematic of generation and maintenance of primary BC cultures in basal cell medium (BCM).
(L) Representative live whole-mount microscopy of P0 and P1 primary BC cultures. Scale bars, 200 μm.
(M) Representative FACS plot of P1 primary BC culture. Samples were analyzed by RT-qPCR in (N), with red sorting gate representing NGFR+ samples and blue sorting gate representing NGFR− samples.
(N) RT-qPCR analysis of P1 primary BC cultures, sorted based on the gates shown in (M). Statistical analysis performed by paired, two-tailed Student’s t test. Bars indicate mean ± SD. n = 3 biological replicates.
(O) Immunofluorescence confocal microscopy of paraffin-embedded tissue sections of primary BC sphere cultures for KRT5, ACTUB, and SCGB1A1 proteins. Nuclei are counterstained with Hoechst33342. Scale bars, 20 μm.
(P) Quantification of cell yield and growth kinetics of primary BC over six serial passages in BCM. Bars indicate mean ± SD. n = 3 biological replicates.
Comparing sorted Nkx2-1mCherry+/NGFR+ cells and sorted Nkx2-1mCherry+/NGFR− cells vs. fresh primary tracheal epithelium, we found expression of canonical BC markers, Krt5 and Trp63, in the NGFR+ population at levels equal to or greater than primary tracheal epithelium, suggesting their identity as bona fide iBCs. In contrast, the smaller population of NGFR− cells were depleted of BC markers and instead expressed airway secretory cell markers, Scgb1a1 and Muc5ac (Figures 1E and 1F). We observed minimal ciliated marker expression (Foxj1) in either PSC-derived population. iBCs could be serially passaged at least 10 times without losing their proliferation potential, generating more than 1015 cells per input cell over 9 passages (Figures 1G and S1B). To assess the stability of canonical gene expression over time, we analyzed sorted NGFR+ and NGFR− iBC cultures at P1, P3, P5, P7, and P9 and found maintained expression of canonical airway epithelial lineage genes, Nkx2-1, Krt5, and Trp63, in the progeny of NGFR+ cells and of Nkx2-1, Scgb1a1, and Muc5ac in NGFR− cells (Figure S1C). Importantly, after cryopreservation and subsequent thawing, iBCs exhibited karyotypic and outgrowth stability in BCM and maintained their Nkx2-1mCherry and BC marker expression profiles, important quality assurance measures required for a freezer-archived source of cells for potential transplant therapies (Figures S1D and S1E).
Next, we characterized the outgrowth of passaged iBCs after extensive serial passaging (P6) using single-cell RNA sequencing (scRNA-seq). Again, we found heterogeneity evident as a mixture of two airway lineages: a major BC-like population (Nkx2-1+, Trp63+, Krt5+) and a minor population of secretory-like cells (Nkx2-1+, Scgb1a1+, Muc5b+). We found little to no evidence of ciliated, alveolar, or non-lung fates emerging (Figures 1H and 1I). The identity of the BC cluster was further confirmed based on the observation that canonical BC markers Krt5, Krt14, Krt17, and Trp63 were all among the top 20 most differentially enriched transcripts in these cells (Figure 1J; Table S1).
To ensure that identical culture conditions could be used to prepare primary BCs for transplantation, we next isolated primary EPCAM+ cells from mouse tracheas for culturing in BCM (Figure 1K). We observed outgrowth of cells expressing BC markers (Figures 1L-1O and S1C) over 5 serial passages and monitored the kinetics of expression of canonical airway epithelial marker genes (Nkx2-1, Krt5, Trp63, Muc5ac, and Foxj1) in sorted NGFR+ vs. NGFR− cells over time. We found evidence of increasing expression of BC markers Krt5 and Trp63 only in the NGFR+ cell fraction (Figure S1C). The proliferation rate of primary BCs slowed at P5, in contrast to iBCs, and we could not expand primary BC beyond P5 (Figure 1P). Taken together, these results indicate successful derivation of mouse iBCs from PSCs and development of a serum-free/feeder-free medium, BCM, that can be used to expand both primary BCs and iBCs in identical culture conditions for comparative transplantation studies.
Airway epithelial reconstitution by mouse primary BC transplantation
Since broadly accepted literature41 indicates that engraftment of hematopoietic stem cells (HSCs) requires bone marrow ablation prior to transplantation in order to make space in the recipient’s stem cell niche, we employed intra-tracheal delivery of polidocanol42 to ablate tracheal airway epithelial cells (Figure S2A). Prior reports have found tracheal epithelial sloughing occurs within 3 h of polidocanol exposure, and by 24 h, surviving proliferating endogenous BCs begin to cover this injured microenvironment.43 We speculated that acute epithelial sloughing might generate a temporarily exposed niche on the airway basement membrane for exogenous cells to land. To determine the time window for BC transplantation post-polidocanol-mediated injury, we analyzed tracheal tissues in a time series following injury. We confirmed extensive epithelial sloughing at 3 h after polidocanol exposure. At 5 h, epithelial sloughing was more complete with sporadic residual individual cells and more denuded basement membrane. By 7 h, residual cells had flattened to cover the basement membrane, and by 12 h, patches of rounded or cuboidal epithelial cells characterized the regenerating epithelium (Figure S2A). Therefore, we elected to transplant donor cells 5 h post-injury.
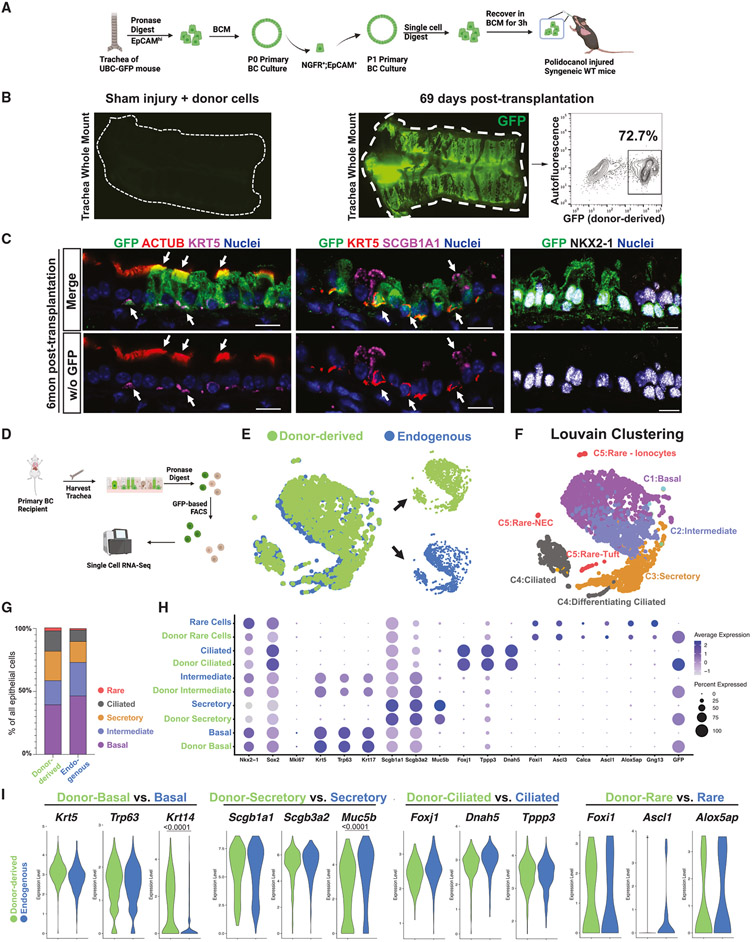
To test the feasibility of engrafting BCs into the tracheal epithelium of injured recipients, we prepared cultured P1 primary BCs obtained from UBC-GFP donor animals and transplanted 6 × 106 cells per animal into immunocompetent syngeneic mice 5 h after polidocanol injury (Figure 2A). As expected, in uninjured recipients (PBS sham injury), we observed no engraftment of transplanted BCs (Figure 2B). In recipients analyzed at 1, 2, and 6 months post-transplantation, we observed GFP+ donor-derived cells retained in recipient injured tracheas. By epifluorescence GFP+ cells contributed to an average of 31.89% ± 18.47% of recipient epithelium (n = 10; Figures 2B, S2B, and S2C), and in some cases, by flow cytometry donor cells comprised as much as 74% of the recipient trachea, indicating significant reconstitution of the tracheal epithelium with donor-derived cells (Figure 2B). By immunofluorescence, we found donor cells localized to the tracheal epithelium, but not detectably to the submucosal glands, and expressed NKX2-1 nuclear protein (Figure 2C), suggesting retention of lung cell fate. Donor-derived cells co-expressed GFP along with markers of trilineage differentiation into airway basal (KRT5+), secretory (SCGB1A1+), or ciliated (ACTUB+) fates (Figure 2C).
Figure 2. Transplantation of mouse primary BCs into syngeneic immunocompetent recipient mice.
(A) Schematic summary of experimental approach for transplantation of cultured primary BC from UBC-GFP mice into polidocanol-injured syngeneic immunocompetent mice.
(B) Tracheal whole-mount fluorescence microscopy and corresponding FACS of GFP (gated from all live cells harvested from the digest) to quantify donor-derived (GFP+) cells present in a representative recipient of primary BCs 69 days post-transplantation. Negative control tracheal whole-mount fluorescence microscopy of a primary recipient exposed to sham (PBS) injury and cell delivery 2 weeks post-transplantation is shown.
(C) Immunofluorescence confocal microscopy for GFP, ACTUB, SCGB1A1, KRT5, and NKX2-1 proteins in a representative primary BC recipient trachea 6 months post-transplantation. White arrows indicate GFP+ (donor-derived) cells co-expressing the indicated canonical airway epithelial markers. Upper panels show all indicated fluorescence channels, and lower panels (“w/o GFP”) have the GFP channel removed. Nuclei are counterstained with Hoechst33342. Scale bars, 10 μm.
(D) Schematic for cell capture and scRNA-seq analysis of endogenous (GFP−) and donor-derived (GFP+) tracheal epithelial cells from recipients of primary BCs.
(E) SPRING visualization of single-cell transcriptomic profiles from primary BC recipient trachea; 69 days post-transplantation. Cells are colored by sample origin. Separated view of donor-derived and endogenous cell clusters are shown (also in Figure S2D).
(F) Louvain clustering and annotation of cells from the SPRING shown in (G) (resolution = 0.2).
(G) Lineage frequency of donor-derived vs. endogenous tracheal epithelial cells.
(H) Dot plot comparing canonical gene expression levels and frequencies between donor-derived cell lineages and their endogenous counterparts.
(I) Violin plot comparing selected canonical gene expression between donor-derived lineages and their endogenous counterparts. Statistical analysis is done with Wilcoxon signed-rank test with adjusted p values shown only for significantly different expression levels.
To further assess the molecular phenotypes of the progeny of transplanted BCs and compare them with their endogenous counterparts in the same recipient, we performed scRNA-seq of 1,807 GFP+ (donor-derived) and 1,294 GFP− (endogenous) epithelial cells from a recipient trachea 69 days post-transplantation. GFP+ and GFP− cells were captured and sequenced simultaneously to avoid batch effects (Figure 2D). We found that the global transcriptomes of donor-derived and endogenous cells were highly similar (Figure 2E, dimensionality reduction visualization of transcriptomes by SPRING44). Louvain clustering analysis identified five cell clusters (Figure 2F), easily distinguished based on expression of known canonical markers of airway epithelial cell types.14,15 Donor-derived cells contributed to all major (basal, secretory, and ciliated) and minor (neuroendocrine, tuft, Hillock, and ionocyte) airway epithelial cell types (Figures 2F-2H and S2E). Both donor-derived and endogenous cells were similarly quiescent, and no alveolar or non-lung fates were detected (Figures 2H and S2E). Donor-derived cells were highly similar to their endogenous counterparts, as evidenced by high Spearman correlation coefficients for each donor vs. endogenous cell lineage (Figures 2H and S3F). For each airway cell type, pairwise comparisons of GFP+ vs. GFP− cells indicated that less than 40 genes were differentially expressed (Table S2). We found no statistically significant difference in canonical lineage marker expression between GFP+ vs. GFP− cells, with the exception of Krt14 for BCs (upregulated in donor-derived) and Scgb3a1 and Muc5b for secretory cells (downregulated in donor-derived, Figure 2I). Together, these results prove the feasibility of engrafting cultured BCs into immunocompetent recipient mouse tracheas after polidocanol-induced epithelial injury.
Airway epithelial reconstitution by mouse iBC transplantation
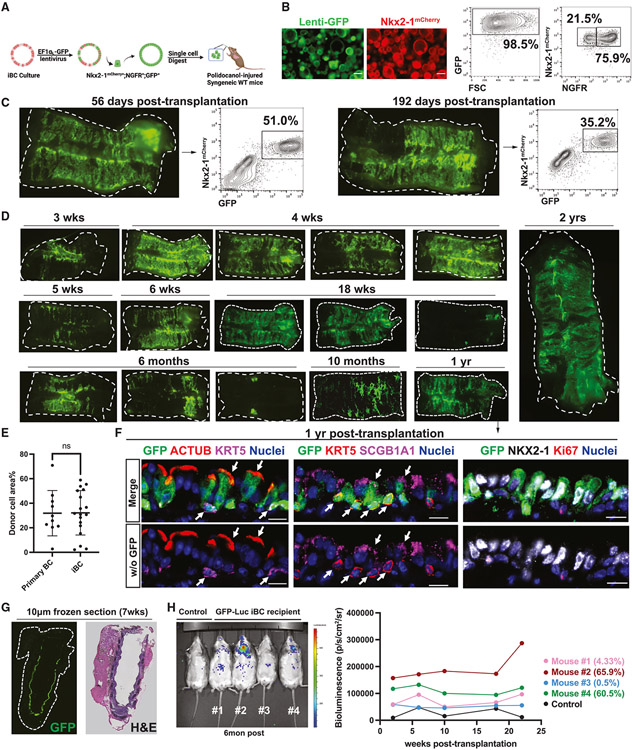
Having established an approach to deliver BCs into the injured mouse trachea, we next sought to test the ability of cultured iBCs to engraft in the stem cell compartment of immunocompetent mouse tracheas. Established cultures of iBCs carrying the Nkx2-1mCherry reporter were labeled with a constitutively GFP-expressing lentivirus and subsequently sorted based on GFP+; Nkx2-1mCherry+;NGFR+ expression to purify GFP-tagged iBCs for further culture expansion or cryopreservation. Following an additional 10–14 days of 3D culture, karyotypically normal (Figure S3A) cells were delivered intratracheally (without further sorting) into syngeneic immunocompetent recipients post-polidocanol injury (Figures 3A and 3B). In recipients analyzed 3 weeks to 2 years post-transplantation (n = 19 total), donor-derived cells were easily detected based on tracheal whole-mount microscopy (Figures 3C and 3D). By epifluorescence quantification, GFP+ cells contributed to an average of 32.19% ± 18.13% of iBC recipients’ tracheal epithelium, similar to the efficiency of primary BC transplants (Figure 3E). In some cases, donor-derived cells comprised more than 50% of the epithelial cells of recipient tracheas by flow cytometry quantitation (Figure 3C). Immunofluorescence studies demonstrated that GFP+ cells retained lung fate (NKX2-1+) and contributed to basal (KRT5+), secretory (SCGB1A1+), and ciliated (ACTUB+) lineages in recipients. E-cadherin immunostaining suggested the expected apicalbasal epithelial polarization of GFP+ cells, and GFP+ BCs were located on the basal epithelial surface next to the basement membrane (Figures 3F and S3B). We did not detect GFP+ cells outside of the epithelial layer or in the submucosal glands (Figure 3G). We found that the majority of recipients exhibited GFP+ cells in the extrapulmonary airways, with a minor fraction extending to intrapulmonary large and small airways (Figure S3C). When donor-derived cells were observed in the intrapulmonary airways, these cells appeared as basal, secretory, and ciliated lineages (Figure S3D).
Figure 3. Transplantation of mouse iBCs into syngeneic immunocompetent recipient mice.
(A) Schematic summary of experimental approach for transplantation of cultured iBC, tagged with lentiviral GFP, into polidocanol-injured syngeneic immunocompetent mice.
(B) Representative whole-mount live fluorescence microscopy and FACS of donor cell population, quantifying NGFR, GFP, Nkx2-1mCherry expression, and forward scatter (FSC). Scale bars, 200 μm.
(C) Whole-mount fluorescence and corresponding FACS quantification (gated from all live cells harvested from the digest) of iBC recipient 56 and 192 days post-transplantation.
(D) Tracheal whole-mount fluorescence microscopy of GFP+ iBCs, shown at various time points (3 weeks to 2 year) post-transplantation.
(E) Quantification of primary BC (n = 10) and iBC (n = 19) transplantation efficiency, calculated by GFP+% area over total recipient trachea area in epifluorescence image. Bars indicate mean ± SD. Comparison between epifluorescence imaging quantification and corresponding FACS transplantation efficiency quantification is shown in Figure S4C.
(F) Immunofluorescence confocal microscopy for GFP, ACTUB, SCGB1A1, KRT5, and NKX2-1 in iBC recipient trachea 1 year post-transplantation. Nuclei are counterstained with Hoechst33342. White arrows indicate GFP+ donor-derived cells co-expressing indicated canonical airway epithelial markers. Lower panels have GFP channel removed. Scale bars, 10 μm.
(G) Tracheal GFP fluorescence in an unfixed, unstained frozen tissue section and H&E staining of the immediately adjacent tissue section of a recipient of GFP+ iBCs, 7 weeks post-transplantation.
(H) In vivo bioluminescence imaging of four recipients and an untransplanted control followed for 6 months after receiving transplanted iBCs tagged with GFP− luciferase-expressing lentivirus. Transplantation efficiency of each recipient (quantified by FACS in Figure S4C) is shown.
Next, we sought to study the dynamics of putative engraftment. We took two approaches: (1) to study initial seeding and outgrowth of transplanted cells, we harvested recipients of GFP-tagged iBCs beginning on the first day post-transplant (dpt) and every 2 days thereafter for the first week, and (2) we tagged iBCs with a luciferase and GFP-expressing lentivirus prior to transplantation, allowing each recipient to be followed longitudinally using intravital bioluminescence imaging. We found that initially (1 dpt), donor-derived basal-like cells (GFP+; KRT5+) represented the majority of the cells, attaching to the basement membrane as single cells and assuming a flattened morphology covering the tracheal basement membrane at sites of injury. By 3 dpt, donor-derived cells formed small patches of KRT5+ cells, with cuboidal secretory differentiation evident by SCGB1A1 immunostaining. At 5 dpt, a reconstituted pseudostratified epithelium arose from contributions of both transplanted and endogenous cells, which was further established by 7 dpt (Figures S4A and S4B). In recipients of luciferase-labeled iBCs (n = 4), followed serially for 22 weeks after transplantation, intravital bioluminescence from transplanted cells was found only in the trachea or lung regions and was stable over time (Figure 3H), with bioluminescence measures of donor cell presence verified by both GFP fluorescence microscopy and flow cytometry at the time of harvest (Figures S4C and S4D). We observed no extrapulmonary bioluminescence signal in any recipient (Figure 3H). Distal lung bioluminescence detected in one of four recipients suggested intrapulmonary engraftment, again consistent with engraftment of donor cells in the intrapulmonary airways of a minor subset of recipients (Figure S3C and S3D).
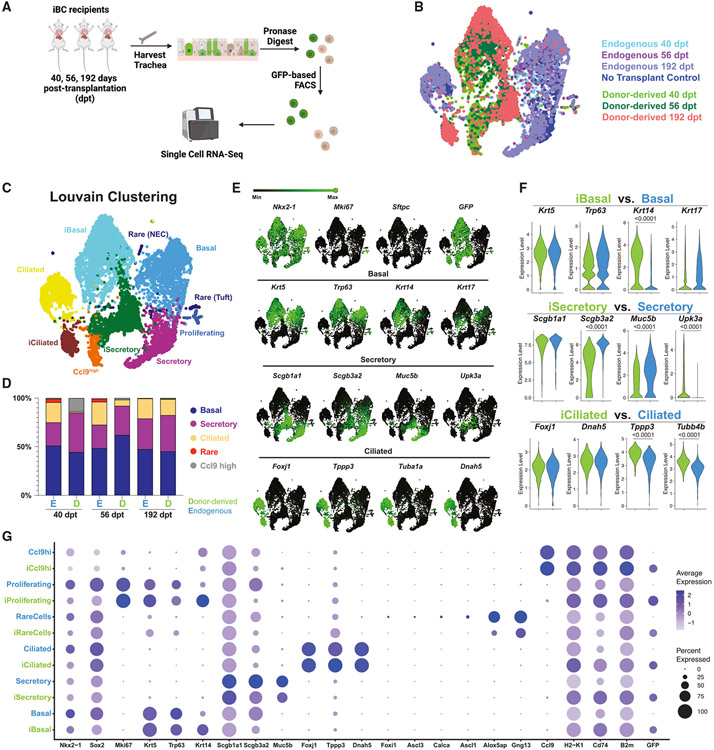
scRNA-seq of iBC-derived cells in vivo
To more completely assess the molecular phenotypes of transplanted iBCs compared with their endogenous counterparts, we performed scRNA-seq of three recipient tracheas after prolonged recovery post-transplantation (40, 56, and 192 dpt), as well as a control trachea that received no injury and no transplantation (Figure 4A). Endogenous cells (GFP−) from each recipient, representing each of the 3 major tracheal epithelial cell lineages (basal, secretory, and ciliated), were evident on dimensionality reduction visualization and expressed global transcriptomes that closely overlapped with uninjured control mouse epithelium, suggesting that transplanted iBCs likely did not significantly perturb the transcriptomes of endogenous cells (Figure 4B). Further, 1,500–1,700 GFP+ cells analyzed per recipient expressed Nkx2-1 and appeared as trilineage differentiated airway populations, each defined based on high levels of expression of canonical-lineage-defining markers for basal (Krt5, Trp63, Krt14), secretory (Scgb1a1, Scgb3a2, Muc5b), and ciliated (Foxj1, Tppp3, Tuba1a) lineages. Louvain clustering of all cells identified nine cell clusters, with GFP+ cells clustering separately from endogenous cells for all three major airway subtypes (Figures 4C-4E). Both donor-derived and endogenous cells were similarly quiescent, and no alveolar or non-lung fates were detected (Figure 4E). Donor-derived rare airway cell types (neuroendocrine, tuft) were detectable as discrete clusters (Figure S5B), but ionocytes (endogenous or donor-derived) were too rare to be reliably identified.
Figure 4. Characterization of iBC donor-derived vs. endogenous epithelial cells in recipient tracheas by single-cell transcriptomic profiling.
(A) Schematic of experimental approach for cell capture and scRNA-seq profiling of epithelial cells from tracheas of three recipients of GFP+ iBCs at 40, 56, and 192 days post-transplantation. One “no transplant” control animal (no transplant, no injury) was included in the experiment as shown in (B).
(B) SPRING visualization of single-cell transcriptomic profiles of tracheal cells from recipients of iBC transplants as shown in (A). Cells are colored either by sample origin.
(C) SPRING visualization of single-cell transcriptomic profiles of tracheal cells from recipients of iBC transplants as shown in (A). Cells are colored by annotated Louvain clustering (resolution = 0.2).
(D) Lineage frequency of donor-derived vs. endogenous tracheal cells for each recipient.
(E) Selected canonical gene expression shown as green overlays on SPRING plot.
(F) Violin plots comparing expression levels of selected canonical marker genes between donor-derived lineages and their endogenous counterparts. Statistical analysis is done with Wilcoxon signed-rank test with adjusted p values shown only for significantly different expression levels.
(G) Dot plot comparing canonical gene expression between donor-derived lineages and their endogenous counterparts.
We noted donor-derived ciliated cells were under-represented, compared with endogenous cells in the two recipients harvested 40 and 56 dpt, but the frequency was more similar in the recipient harvested at 192 dpt (Figure 4D). In addition, in 2/3 recipients, we detected a distinct population of donor-derived Ccl9high cells, clustering adjacent to secretory club cells, which is enriched in H2-K1 and lncRNA AW112010, two markers recently reported to distinguish in vivo rare “club-like” progenitors that possess lung epithelial regenerative capacity.24 Like their published counterparts, these cells were CD74high and Scgb1a1low (Figure S5C). Gene set enrichment analysis revealed that immune responses and antigen presentations were among the most enriched ontologies in these Ccl9high cells based on 1,406 differentially expressed genes in this cluster (Figure S5D; Table S3), similar to the published findings on the H2-K1high progenitors.24
Next, we performed pairwise comparisons between the transcriptomes of donor-derived vs. endogenous cells for each of the three major airway cell types. We found similar levels of expression of canonical-lineage-defining transcripts (e.g., Nkx2-1, Trp63, Scgb1a1, and Foxj1; Figures 4E-4G). Across all lineages in donor-derived cells, we found transcripts encoding major histocompatibility complex (MHC) genes such as H2-Eb1, B2m, and Cd74 among the top 20 differentially expressed genes (Figure S5E; Table S4). Analyzing each lineage separately, we found donor-derived BCs had higher expression of Krt14 and a trend of lower Krt17 (Figures 4E, 4F, and S5E); donor-derived secretory cells had significant gene expression differences in proximal-distal patterning markers, compared with endogenous cells. For example, donor-derived secretory cells expressed significantly higher levels of distal secretory marker Upk3a45 and significantly lower levels of proximal secretory markers Reg3g, Cbr2, Chad, and Scgb3a2,45 compared with endogenous secretory cells (Figures 4E, 4F, and S5E-S5H). Donor-derived ciliated cells expressed higher levels of Tubb4b but lower levels of cilia regulatory genes such as Drc1 (Figures 4F and S5E).
Function of donor-derived cells
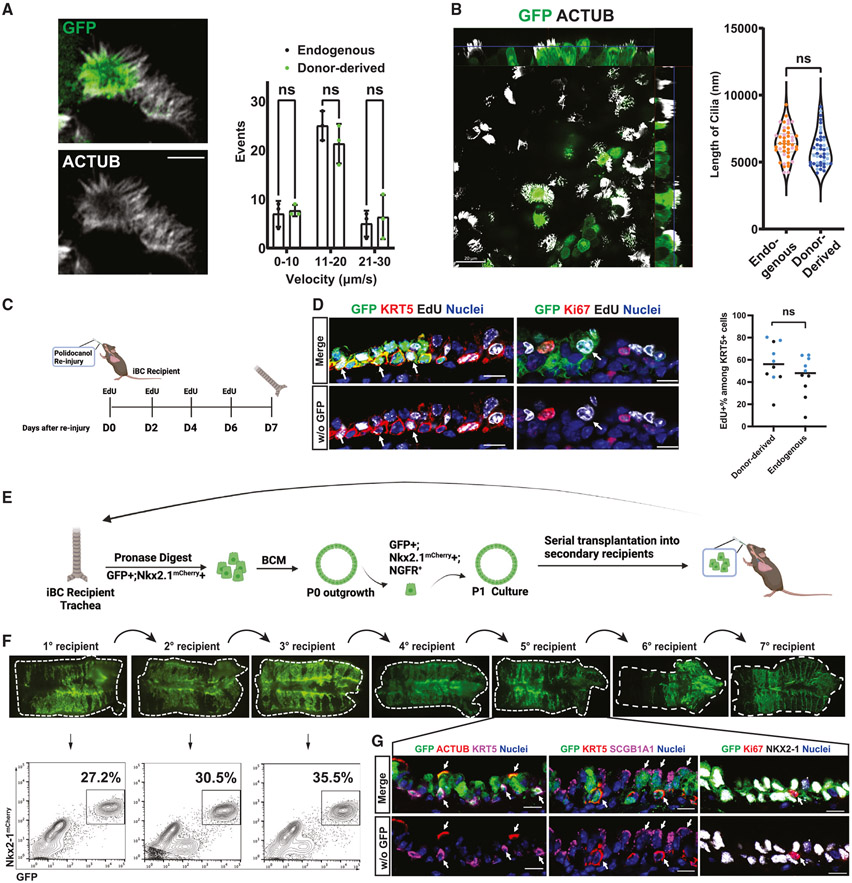
Next, we sought to characterize the in vivo function of donor-derived cells. We first quantified the morphology and function of motile cilia in donor-derived vs. endogenous ciliated cells. Using freshly dissected trachea, motile cilia were readily observed by high-resolution microscopy (Video S1). Immunocytochemistry of motile cilia showed no gross morphological differences between the iBC-derived ciliated cells and endogenous ciliated cells (Figure 5A). Kymograph analysis revealed no statistically significant difference in beating frequencies between donor-derived and endogenous cilia (Figure 5A). We also measured ciliary length from fixed tracheal tissue and found no significant difference in cilia length between donor-derived and endogenous ciliated cells (Figure 5B).
Figure 5. In vivo stem cell self-renewal and differentiated cell function of engrafted murine iBC-derived cells.
(A) Representative screen capture of live GFP fluorescence by high content video confocal microscopy imaging, indicating GFP+ (donor-derived) and GFP− (endogenous) multiciliated cells (cilia labeled with wheat germ agglutinin in white); graph shows quantification of cilia beating frequency (events per unit time) of donor-derived and endogenous cells. Bars indicate mean ± SD. Scale bars, 5 μm.
(B) Whole-mount immunofluorescence microscopy of GFP and acetylated tubulin (ACTUB) proteins in recipient trachea 310 days after transplantation of GFP+ iBCs, indicating donor-derived (GFP+) multiciliated cells; graph shows quantification of cilia length in donor-derived and endogenous multiciliated cells. n = 44 cells per condition. Scale bars, 20 μm.
(C) Schematic summary of experimental approach for assessing response of donor iBC-derived engrafted cells to in vivo re-injury from repeated polidocanol exposure.
(D) Immunofluorescence confocal microscopy of representative tracheal tissue section for GFP, KRT5, EdU, and Ki67 in iBC recipient trachea 1 week post-second polidocanol injury and corresponding quantification. Nuclei are counterstained with Hoechst33342. White arrows indicate donor-derived (GFP+) cells that co-label with EdU. Lower panels have GFP channel removed. Scale bars, 10 μm. n = 2 animals (411 donor-derived KRT5+ cells and 262 endogenous KRT5+ cells). Statistical analysis performed by unpaired, two-tailed Student’s t test.
(E) Schematic summary of experimental approach for serial transplantations of GFP+-tagged iBCs into secondary recipients to measure self-renewal capacity.
(F) Tracheal whole-mount fluorescence microscopy and corresponding FACS analysis (gated from all live cells harvested from the digest) of secondary recipients of GFP+ iBCs, characterizing seven generations of serial iBC transplants. 7th generation (7°) recipient trachea was also shown in analysis in Figure S6A.
(G) Immunofluorescence confocal microscopy for GFP, ACTUB, SCGB1A1, KRT5, and NKX2-1 in a 5th generation (5°) iBC recipient trachea. Nuclei are counterstained with Hoechst33342. White arrows indicate GFP+ donor-derived cells co-expressing indicated canonical airway epithelial markers. Lower panel have GFP channel removed. Scale bars, 10 μm.
Next, we tested whether donor-derived BCs can participate in post-injury repair in response to a second insult in vivo following engraftment. We performed a second round of polidocanol injury in iBC recipients 6 weeks post-transplantation and labeled all proliferating cells in vivo with 5-ethynyl-2’-deoxyuridine (EdU) pulses for 1 week post-injury (Figure 5C). Immunofluorescence microscopy demonstrated GFP+/EdU+/KRT5+ tracheal epithelial cells, indicating engrafted iBCs can re-enter cell cycle and proliferate in response to injury. In addition, we also observed GFP+/EdU+/Ki67− tracheal epithelial cells (Figure 5D), suggesting donor-derived cells that had proliferated in the first week after re-injury (during EdU pulse) were able to resume quiescence during the EdU chase period. Importantly, the percentage of EdU+ donor-derived cells was not different from that of endogenous cells, suggesting similar regenerative potential (Figure 5D).
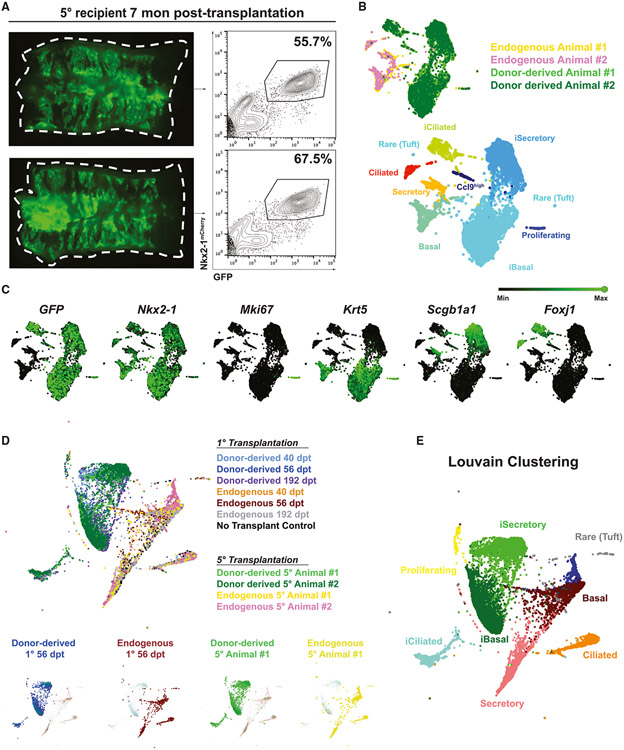
To evaluate whether the key stem cell functions of self-renewal and differentiation had been reconstituted with donor-derived cells, we next evaluated the capacity of engrafted iBCs to contribute to other recipients through serial transplantations. Murine HSCs of the bone marrow have been shown to reconstitute multilineage blood lineages after six generations of serial transplantation in myeloablated mice.46 To recapitulate this gold standard assay of stem cell self-renewal, we harvested GFP+ donor-derived cells 6 weeks post-transplantation, expanded them in BCM to generate the cell number needed for transplantation, and transplanted the donor-derived cells into secondary polidocanol-injured syngeneic recipients (Figure 5E). Through multiple generations of serial transplantations, we observed robust GFP+ tracheal epithelial repopulating capacity, undiminished even after seven generations of transplantation (Figures 5F and 5G). By immunofluorescence, donor-derived cells retained trilineage differentiation potential in vivo (Figure 5H). To investigate whether the transcriptomic profile of the donor-derived cells changed over the generations of secondary transplantations, we performed scRNA-seq of the tracheal epithelium of two fifth-generation recipients (Figure 6A). We found that the donor-derived cells, while contributing to all three airway epithelial lineages in the fifth-generation recipients, still clustered separately from their endogenous counterparts (Figures 6B and 6C). Integrating this dataset with that of the first-generation recipients shown in Figure 4 without harmonization, we found that all donor-derived cells clustered together and were indistinguishable by Louvain clustering (Figures 6D and 6E), suggesting a stably distinct transcriptomic profile of donor-derived cells over time. In addition, to test whether the regenerative potential of iBCs exhaust over time, we performed a time-series experiment, harvesting 7th generation serial transplantation recipients at 1, 3, 5, or 7 dpt. Similar to our findings in primary (first-generation) recipients (Figure S4), the donor-derived cells were again able to generate small colonies and differentiate into secretory cell lineages at 3 dpt and form pseudostratified epithelium at 5 dpt, which were further established by 7 dpt (Figure S6), suggesting maintenance of regenerative potential. In total more than 20 months elapsed between first transplantation and final harvest of the last transplanted generation of serial recipients, indicating both the longevity of the iBCs as well as their self-renewal capacity.
Figure 6. Characterization of iBC donor-derived vs. endogenous epithelial cells in 5th generation recipient tracheas by single-cell transcriptomic profiling.
(A) Whole-mount epifluorescence imaging and corresponding FACS analysis of 5° generation recipient animals profiled by scRNA-seq.
(B) SPRING visualization of scRNA-seq of 5th generation recipients, colored by sample or by Louvain clustering (resolution = 0.2).
(C) Selected canonical gene expression shown as green overlays on SPRING plot.
(D) SPRING visualization of merged scRNA-seq dataset of 1° recipients (Figure 4) and 5° recipients without harmonization. Selected samples are highlighted in the lower small panels.
(E) SPRING visualization of merged scRNA-seq dataset of 1° recipients (Figure 4) and 5° recipients without harmonization, colored by Louvain clustering (resolution = 0.2).
Airway epithelial reconstitution by the transplantation of human primary BCs or iBCs
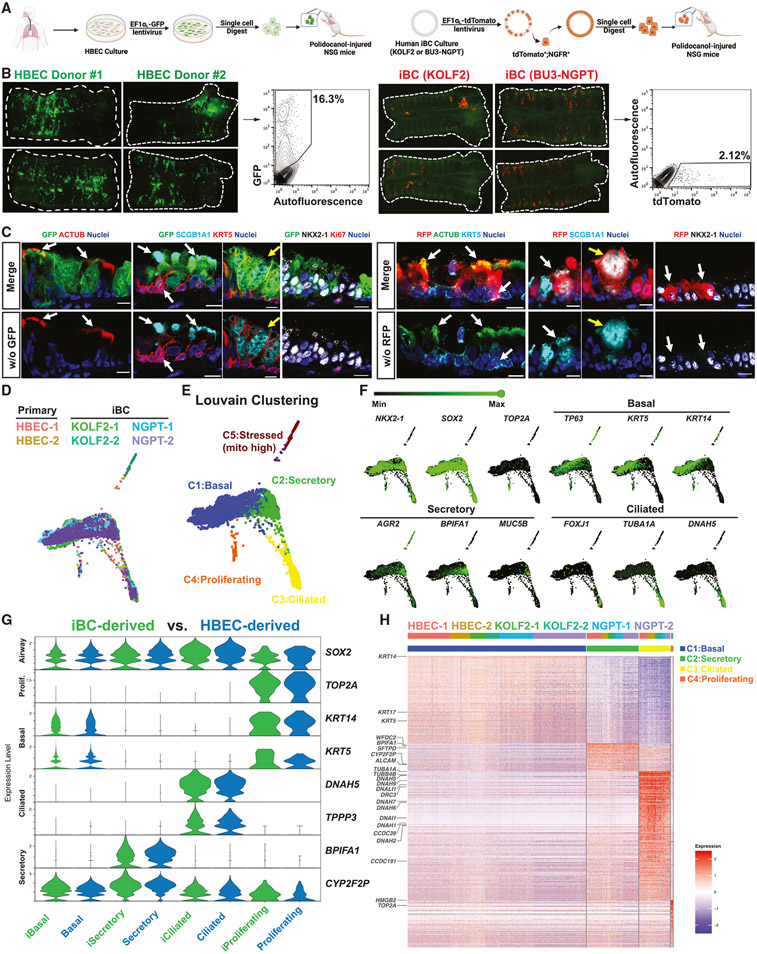
Having established the airway transplantation approach with mouse BCs and iBCs, we next sought to further our preclinical model by transplanting human primary BCs (human bronchial epithelial cells, HBECs) and human iBCs.29 For HBEC transplantation, we thawed cryopreserved P1 HBECs from two different donors and tagged the cells with a GFP-expressing lentivirus before transplantation into NSG recipient mice following polidocanol tracheal injury (Figure 7A). For human iBC transplantation, we prepared 3D cultures of two human iBC lines of different genetic backgrounds (KOLF247 and BU3-NGPT29) and tagged the cells with a tdTomato-expressing lentiviral vector. We purified tdTomato+/NGFR+ iBCs and transplanted these cells into NSG recipient animals (Figure 7A). We detected donor-derived cells from both HBECs and iBCs in recipient tracheas 6 weeks post-transplantation by whole-mount microscopy and flow cytometry (Figures 7B and S7A). HBEC donors (n = 4) contributed to 15.97% ± 1.14% of recipients’ tracheal epithelium, while human iBCs (KOLF2 n = 5; BU3-NGPT n = 11) contributed to 1.831% ± 1.327% (Figure S7B). Immunofluorescence indicated that human donor-derived cells retained lung fate (NKX2-1+) and contributed to basal (KRT5+), secretory (SCGB1A1+), and ciliated (ACTUB+) lineages in each recipient (Figure 7C). Interestingly, we found SCGB1A1+ cells of goblet-like morphology (Figure 7C, yellow arrows), which are normally found in human, but not mouse, tracheas. Because human cell xeno-transplantation into NSG mice resulted in markedly fewer engrafted cells, compared with our prior syngeneic mouse cell into immunocompetent mice transplants, we considered whether this was due to the recipient strain. We thus transplanted murine GFP+ iBCs into NSG recipients and observed GFP+ airway epithelial trilineage progeny at 18 months post-transplantation, with similar efficiencies to what had been observed in immunocompetent recipients (Figures S7C-S7E).
Figure 7. Characterization of putative engrafted human primary BCs or iBCs in injured NSG mouse recipient tracheas.
(A) Schematic of transplantation of human primary BCs (human bronchial epithelial cells, HBECs) or iBCs into polidocanol-injured NSG mouse trachea. Note lentiviral GFP or tdTomato tagging of cultured cells prior to transplantation.
(B) Whole-mount fluorescence and corresponding FACS quantification (gated from all live cells harvested from the digest) of derivatives of either GFP+ human primary BCs or tdTomato+ human iBCs in mouse recipient tracheas 6 weeks post-transplantation.
(C) Immunofluorescence confocal microscopy for GFP, tdTomato (RFP), ACTUB, SCGB1A1, KRT5, and NKX2-1 in mouse recipient tracheas 6 weeks post-transplantation of human cells. Nuclei are counterstained with Hoechst33342. White arrows indicate GFP+ or RFP+ human donor-derived cells co-expressing canonical airway epithelial markers. Yellow arrows indicate human donor-derived cells with goblet cell morphology. Lower panels have GFP or RFP channel removed, as indicated. Scale bars, 10 μm.
(D) Single-cell transcriptomic profiling of human donor-derived cells in recipient mouse tracheas; SPRING visualization representing scRNA-seq of human cells in mouse recipient tracheas profiled 6 weeks post-transplantation. Cells are colored by sample origin.
(E) Louvain clustering and annotations of human cells in each recipient from (D) (resolution = 0.1).
(F) Selected canonical marker gene expression (green) overlayed on SPRING plots.
(G) Violin plots comparing selected canonical gene expression levels between putatively engrafted human primary BC-derived lineages vs. human iBC-derived lineages.
(H) Heatmap of the top 532 most variable genes (filtered by logFC > 0.5 and FDR-adjusted p < 0.05; across Louvain clusters from (E). Stressed population (C5) is excluded from this heatmap analysis.
Given that human BCs extend more distally in the respiratory tree, compared with their murine counterparts, we next studied the intrapulmonary airways of recipients. We observed GFP+ HBEC-derived ciliated and secretory cells comprising a columnar epithelium that was taller than the adjacent murine host epithelium (Figures S7F and S7G). Notably, human donor-derived KRT5+ BCs and MUC5B+ goblet-like cells, which are not normally present in mouse intrapulmonary airways, were detected (Figure S7G). Human iBCs, in contrast, contributed mostly to the tracheal epithelium with only rare cells found in the intrapulmonary airways (Figure S7F).
To more extensively characterize the molecular phenotypes of the transplanted human cells and compare transplanted HBECs with human iBCs, we performed scRNA-seq on GFP+ or tdTomato+ cells isolated from six recipient animal tracheas (two HBEC Donor#2 recipients, two KOLF2 iBC recipients, and two BU3-NGPT iBC recipients) 6 weeks post-transplantation. Human single-cell transcriptomes of putative engrafted cells, visualized by SPRING, overlapped across all six recipients without any data harmonization, suggesting a high level of similarity regardless of origin (Figure 7D). Louvain clustering revealed five main clusters, annotated based on canonical-lineage-defining markers as basal (KRT5, TP63), ciliated (FOXJ1, DNAH5), secretory (AGR2, MUC5B), proliferating (TOP2A), and a minor stressed population with high mitochondrial gene percentage (Figure 7E, 7F, and S7H). All human samples contributed to all major clusters (Figures 7H and S7H). Donor-derived cells from HBECs and iBCs were highly similar to each other in all three lineages based on canonical gene expression and were not distinguishable by Louvain clustering (Figures 7G and 7H; Table S5).
DISCUSSION
Our results demonstrate in vivo airway epithelial engraftment of mouse and human primary BCs or iBCs. Using directed differentiation of PSCs to prepare exogenous airway basal-like cells for transplantation provided an easily accessible alternative to primary cells, which sometimes can be challenging to obtain or expand from patients. Importantly, transplantation of BCs, whether primary or engineered from PSCs, resulted in reconstitution of the endogenous basal stem cell compartment of the pulmonary airways, with engrafted cells capable of long-term self-renewal and durable multipotent differentiation capacity for at least 2 years in vivo.
Reconstitution of a tissue-resident stem cell compartment with primary and engineered cells at high efficiency in an immunocompetent host has considerable implications for regenerative therapy. Our findings extend those of prior reports of airway epithelial transplantation,21-28 where immunodeficient recipient animals displaying evidence of grafted epithelial cells were followed for shorter periods with less formal tests of in vivo stem cell functional reconstitution. We found mouse primary BCs and iBCs, when transplanted into syngeneic recipients without immunosuppression, can replace more than 50% of the endogenous tracheal epithelium after a single treatment. This degree of engraftment efficiency, if performed with gene-corrected cells, is likely to provide sufficient functional replacement of endogenous cells compromised by disease-driving gene mutations. Engraftment of immunocompetent recipients with syngeneic cells also simulates potential future autologous cell transplantation approaches that can be adapted to reconstitute the airways of individuals with cystic fibrosis or primary ciliary dyskinesia, using their own induced pluripotent stem cell (iPSC)-derived BCs after in vitro gene editing.
One major hurdle limiting cell transplantation is producing large numbers of donor cells likely to be needed for the procedure. Airway BCs are the resident stem cells of the mouse trachea and human pseudostratified airway epithelium,6,14,15 but obtaining these cells in sufficient quantity from patients already compromised by lung disease can be challenging. Primary human bronchial cells harvested via bronchoscopy from some patients or from explant lung tissues from others offer one source of cultured BCs for transplantation, as used in our studies; however, some potential hurdles that might limit access to this resource for some patients include the following: (1) the invasive procedures required to harvest these cells48,49; (2) potential loss of BC phenotype over serially passaged cultures50,51; and (3) retained epigenetic signatures of disease in primary cells.52 Our findings suggest iBCs as an alternative source of cells for transplantation that are easily accessible, archivable,29 readily gene edited to correct disease-causing mutations,29,53 and have been reprogrammed to erase epigenetic disease-associated signatures. Importantly, our human transplantation model offers a platform that can be employed to test and compare the in vivo functions, reconstituting potential, and theoretical advantages and disadvantages of both primary and engineered human basal-like cells. The similarity of the single-cell transcriptomes of the progeny deriving in vivo from each human cell type after transplantation implies either cell type may be used for airway engraftment, at least in the case of normal BCs and iBCs, and future work will now need to focus on how cells from individuals with lung disease might function in vivo after engraftment. In addition, long-term studies will need to focus on the safety of the transplanted cells, especially screening for potential malignant transformation. In our studies, we did not observe any tumor formation from donor-derived cells in any recipient animal, including in NSG recipients that are more prone to tumor formation.
A key issue to address in transplantation of exogenously cultured cells is their similarity or difference from their endogenous in vivo counterparts, particularly for PSC-derived lineages that have been reported to be less mature than primary lineages in some cases.54-59 Our transplanted mouse primary BCs contributed to major (basal, secretory, ciliated) and rare (ionocyte, neuroendocrine, tuft, hillock) airway epithelial lineages that were almost indistinguishable from their endogenous counterparts according to transcriptomic analyses. In contrast, iBC-derived cells, while contributing to all major airway epithelial lineages with similar expression of most canonical marker transcripts, remained distinct from their corresponding endogenous lineages in terms of their global transcriptional profiles. We found that both iBC-derived basal and secretory lineages exhibited differences in regional patterning. For example, iBC-derived secretory cells exhibited equal Scgb1a1 transcript expression to endogenous tracheal secretory cells but lower Reg3g, Muc5b, Scgb3a2, Chad, and Cbr2, potentially indicating a less proximal (tracheal) molecular phenotype based on previously reported markers of club cell proximal-distal patterning.45 Some of these differentially expressed genes have also been recently reported as markers of a proximally localized club cell subpopulation.60 One potential explanation for such differences is that iBCs may be capable of giving rise to epithelial lineages representing molecular phenotypes of both proximal and distal airways despite their engrafted location in the trachea. Further work will be needed to test this hypothesis, including comparative profiling with intrapulmonary endogenous lineages. Also requiring further study are the notable donor-derived intrapulmonary BCs observed in some recipients of mouse iBCs or HBECs. Since intrapulmonary BCs in mouse airways are infrequently observed, except in settings of injury, future work will be needed to determine the mechanisms and putative stem cell niches responsible for these persistent intrapulmonary airway phenotypes. The derivation from human donor cells of intrapulmonary tall columnar goblet cells, a phenotype also not normally found in the murine airways, also warrants further study as this finding implies this fate is determined by the cell of origin in our studies.
Importantly, despite their transcriptomic differences, mouse iBC-derived lineages exhibited normal in vivo function, such as motile cilia beating from multiciliated cells and post-injury proliferation from BCs. Interestingly, in contrast to mouse iBC derivatives, human primary and iBC-derived lineages across all three different genetic backgrounds showed high levels of transcriptomic similarity in vivo after engraftment.
In summary, our results establish airway stem cell transplantation with exogenous mouse and human primary or PSC-derived basal stem cells to durably reconstitute the airway stem cell compartment in vivo. Our study may thus serve as a first step toward future cell-based therapies, whereby autologous gene-corrected cells may eventually be employed for in vivo reconstitution of airway epithelial function for individuals with genetic airway diseases.
Limitations
While this work establishes the feasibility of transplantation of primary and PSC-derived cells into mouse trachea, future studies will be required to transition into more clinically relevant approaches, particularly with regard to epithelial conditioning regimens. Our transplantation model requires pre-exposure to polidocanol to injure the murine tracheal epithelium, an approach that is unlikely to be feasible in clinical practice. Efforts are underway to develop more clinically relevant conditioning regimens, including localized airway epithelial denudation with detergents,61 and ex vivo perfusion systems to maintain organ function in recipients of airway-instilled cells,61 which may facilitate clinical adaptation of our approach. In addition, localized delivery technologies such as bronchoscopy are likely to be important to selectively graft locally prepared lobes or intrapulmonary airways in the future.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Darrell Kotton (dkotton@bu.edu).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact without restriction.
Data and code availability
Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. The sequencing data will also be available on the Kotton Lab’s Bioinformatics Portal at https://www.kottonlab.com. This paper does not report original code. Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit Anti-NGFR Antibody (no longer in production) | Abcam | Cat#ab8875; RRID:AB_306828 |
| Mouse Anti-NGFR Antibody | Abcam | Cat#ab245134 |
| BV421 Rat Anti-Mouse CD326 | BD Biosciences | Cat#563214; RRID:AB_2738073 |
| Goat Anti-GFP Antibody | US Biological | Cat#G8965-01E |
| Chicken Anti-GFP Antibody | Aves Labs | Cat#GFP-1020; RRID:AB_10000240 |
| Goat Anti-RFP Antibody | My Biosource | Cat#MBS448122 |
| Rat Anti-Mki67 Antibody | Thermo Fisher | Cat#14-5698-82; RRID:AB_10854564 |
| Rabbit Anti-NKX2-1 Antibody | Abcam | Cat#ab76013; RRID:AB_1310784 |
| Rabbit Anti-KRT5 Antibody | BioLegend | Cat#905501; RRID:AB_2565050 |
| Rabbit Anti-RFP Antibody | Rockland | Cat#600-401-379; RRID:AB_2209751 |
| Mouse Anti-Acetylated-Tubulin Antibody | Sigma | Cat#T7451; RRID:AB_609894 |
| Goat Anti-SCGB1A1 (mouse) Antibody | Gift from Barry Stripp | N/A |
| Goat Anti-SCGB1A1 (human) Antibody | Sigma | Cat#ABS1673; RRID:AB_2910611 |
| Rat Anti-E-Cadherin Antibody | Invitrogen | Cat#13-1900; RRID:AB_86571 |
| Alexa Fluor 488 donkey anti-chicken IgY (IgG) (H+L) | Jackson Immuno Research | Cat#703-545-155; RRID:AB_2340375 |
| Alexa Fluor 488 donkey anti-goat IgG (H+L) | Jackson Immuno Research | Cat#705-545-003; RRID:AB_2340428 |
| Alexa Fluor 488 donkey anti-rat IgG (H+L) | Jackson Immuno Research | Cat#712-546-153; RRID:AB_2340686 |
| Alexa Fluor 488 donkey anti-rabbit IgG (H+L) | Jackson Immuno Research | Cat#711-546-152; RRID:AB_2340619 |
| Alexa Fluor 488 donkey anti-mouse IgG (H+L) | Jackson Immuno Research | Cat#715-545-150; RRID:AB_2340846 |
| Alexa Fluor 546 donkey anti-Rabbit IgG (H+L) | Thermo Fisher | Cat#A11055; RRID:AB_2534102 |
| Alexa Fluor 568 donkey anti-mouse IgG (H+L) | Thermo Fisher | Cat#A10037; RRID:AB_2534013 |
| Cy3 donkey anti-rat IgG (H+L) | Jackson Immuno Research | Cat#712-166-153; RRID:AB_2340669 |
| Cy3 donkey anti-rabbit IgG (H+L) | Jackson Immuno Research | Cat#711-165-152; RRID:AB_2307443 |
| Cy3 donkey anti-goat IgG (H+L) | Jackson Immuno Research | Cat#705-165-147; RRID:AB_2307351 |
| Alexa Fluor 647 donkey anti-rabbit IgG (H+L) | Thermo Fisher | Cat#A32795; RRID:AB_2762835 |
| Alexa Fluor 647 donkey anti-goat IgG (H+L) | Jackson Immuno Research | Cat#705-605-003; RRID:AB_2340436 |
| Bacterial and virus strains | ||
| pHAGE-EF1αL-GFP-W lentivirus | Darrell Kotton Lab | Addgene126686 |
| pHAGE-EF1αL-tdTomato-W lentivirus | Darrell Kotton Lab | N/A |
| CMV-Luciferase-EF1α-copGFP lentivirus | System Biosciences | Cat#BLIV511PA-1 |
| Biological samples | ||
| Primary human airway epithelial cell culture | The MLI Tissue Procurement and Cell Culture Core, University of North Carolina, Chapel Hill | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| DMEM | Gibco | Cat#2414671 |
| IMDM | ThermoFisher | Cat#12440053 |
| Glutamax | Life Technologies | Cat#35050-061 |
| Ham’s F12 | Cellgro | Cat#10-080-CV |
| B27 supplement | Invitrogen | Cat#17504-44 |
| N2 Supplement | Invitrogen | Cat#17502-048 |
| Primocin | Invivogen | Cat#ANTPM1 |
| 7.5% BSA Fraction V | ThermoFisher | Cat#15260-037 |
| 1-thioglycerol (MTG) | Sigma | Cat#M6145 |
| Ascorbic Acid | Sigma | Cat#A4544 |
| Recombinant human/mouse/rat Activin A | R&D Systems | Cat#338-AC |
| Recombinant mouse Wnt3a | R&D Systems | Cat#1324-WN-010 |
| Recombinant human Bmp4 | R&D Systems | Cat#314-BP |
| Recombinant mouse Noggin | R&D Systems | Cat#1967-NG |
| SB431542 | Sigma | Cat#S4317 |
| Recombinant human FGF2 | R&D Systems | Cat#233-FB |
| Recombinant human FGF10 | R&D Systems | Cat#345-FG |
| DMH-1 | Tocris | Cat#4126 |
| A8301 | Tocris | Cat#2939 |
| Y-27632 (ROCK inhibitor) | Tocris | Cat#1254 |
| Heparin Salt | Sigma Millipore | Cat#H3393 |
| Growth Factor Reduced 3D matrigel | Corning | Cat#354230 |
| Dispase | Gibco | Cat#17105-041 |
| Collagenase Type IV | ThermoFisher | Cat#17104019 |
| Pneumacault ExPlus Medium | Stem Cell Technologies | Cat#5040 |
| BEGM | The MLI Tissue Procurement and Cell Culture Core, University of North Carolina, Chapel Hill | N/A |
| Retinoic acid | Sigma | Cat#R2625 |
| 0.05% Trypsin-EDTA | Gibco | Cat#25300062 |
| fetal bovine serum | Gibco | Cat#16141079 |
| PureCol | Advanced Biomatrix | Cat#5005 |
| Paraformaldehyde | Electron Microscopy Sciences | Cat#19208 |
| Normal Donkey Serum | Sigma | Cat#D9663 |
| Antigen Unmasking Solution, Citric Acid Based | Vector Laboratories | Cat#H-3300-250 |
| M.O.M. Immunodetection Kit | Vector Laboratories | Cat#BMK-2202 |
| Bovine Serum Albumin | Fisher | Cat#BP1600 |
| FluoroSave Mounting Agent | Millipore | Cat#345789 |
| Hoecsht 33342 | Thermo Fisher | Cat#H3570 |
| Triton-X | Sigma | Cat#T9284 |
| Pronase | Roche | Cat#10165921001 |
| Wheat Germ Agglutinin, Alexa Fluor™ 647 Conjugate | ThermoFisher | Cat#W32466 |
| DNAse | Roche | Cat#10104159001 |
| Polidocanol | Sigma | Cat#88315 |
| XenoLight D-Luciferin | PerkinElmer | Cat#122799 |
| Calcein Blue | ThermoFisher | Cat#C1429 |
| DRAQ7 | Abcam | Cat#ab109202 |
| Critical commercial assays | ||
| RNeasy Mini Kit | QIAGEN | Cat#74104 |
| QIAzol Lysis Reagent QIAGEN | QIAGEN | Cat#79306 |
| RLT Plus lysis buffer | QIAGEN | Cat#1053393 |
| TaqMan Fast Universal PCR Master Mix (2X) | Thermo Fisher | Cat#4364103 |
| High-Capacity cDNA Reverse Transcription Kit | Applied Biosystems | Cat#4368814 |
| Click-iT EdU Cell Proliferation Kit | ThermoFisher | Cat#C10340 |
| Deposited data | ||
| Sequence Data | This Paper | GEO: Super Series GSE211998 |
| Experimental models: Cell lines | ||
| Nkx2-1mCherry mouse ES Cell Line | Bilodeau et al.37 | N/A |
| Human BU3-NGPT iPSC Cell Line | Hawkins et al.29 | N/A |
| Human KOLF2 iPSC Cell Line | Skarnes et al.47 | N/A |
| Experimental models: Organisms/strains | ||
| UBC-GFP mice | Jackson Labs | JAX004353 |
| C57BL/6J mice | Jackson Labs | JAX000664 |
| 129X1/SvJ mice | Jackson Labs | JAX000691 |
| 129S1/SvlmJ mice | Jackson Labs | JAX002448 |
| NSG mice | Jackson Labs | JAX005557 |
| Software and algorithms | ||
| FlowJo Software v.10.8.1 | Becton Dickinson & Company | https://www.flowjo.com/solutions/flowjo |
| Seurat v.3 | Sajita Lab65 | https://github.com/satijalab/seurat |
| SRPING | Klein Lab44 | https://github.com/AllonKleinLab/SPRING_dev |
| Living Image Software v. 3.2 | Caliper LifeSciences | https://www.perkinelmer.com/product/spectrum-200-living-image-v4series-1-128113 |
| Graphpad Prism v.9.5.1 | GraphPad Software | https://www.graphpad.com/features |
| ImageJ v.2.1.0/1.53i | NIH | https://imagej.net/ij/index.html |
| Other | ||
| Taqman Probe: Nkx2-1 | Thermo Fisher | Mm00447558_m1 |
| Taqman Probe: Trp63 | Thermo Fisher | Mm00495788_m1 |
| Taqman Probe: Krt5 | Thermo Fisher | Mm01305291_g1 |
| Taqman Probe: Ngfr | Thermo Fisher | Mm00446296_m1 |
| Taqman Probe: Scgb1a1 | Thermo Fisher | Mm00442046_m1 |
| Taqman Probe: Muc5ac | Thermo Fisher | Mm01276718_m1 |
| Taqman Probe: Foxj1 | Thermo Fisher | Mm01267279_m1 |
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Animals
All studies involving mice were approved by the Institutional Animal Care and Use Committee of Boston University School of Medicine. All transplantation experiments were performed on 8-16 week old adult male mice, as follows. For primary BC transplantations, donor cells were harvested from 8-16 weeks old male C57BL/6J background UBC-GFP transgenic mice animals (JAX004353). Recipient WT immunocompetent C57BL/6J mice were purchased from Jackson Laboratory (JAX000664). For murine iBC transplantations, WT immunocompetent recipient mice were F1 offspring generated in house by crossing 129S1/SvlmJ Males (JAX002448) and 129X1/SvJ Females (JAX000691). For human primary and iBC transplantation experiments, 10-12 week old NSG recipient mice (JAX005557) were used.
Cell lines
Primary murine basal cell culture was established with harvested airway basal cells from C57BL/6J animals. Mouse iBCs were generated from a previously published37,38 Nkx2.1mCherry reporter R1 ESC line (129X1SvJ x 129S1/Sv background).
Primary human bronchial epithelial cells (HBEC) were generated from lung tissue of deceased donors. Human iBCs were generated from previously reported KOLF247 and BU3 NGPT29 human iPSCs.
The culture conditions for different cell lines are detailed in the “methods details” section below.
METHODS DETAILS
Generation and maintenance of murine iBCs
Cell differentiation was performed in a complete serum free differentiation medium (cSFDM) consisting of a base medium of IMDM (ThermoFisher, 12440053) and Ham’s F12 (Cellgro, 10-080-CV) with B27 Supplement with retinoic acid (Invitrogen, 17504-44), N2 Supplement (Invitrogen, 17502-048), 0.1% bovine serum albumin Fraction V (Life Technologies, 15260-037), monothioglycerol (Sigma, M6145), Glutamax (Life Techologies, 35050-061), ascorbic acid (Sigma, A4544), and Primocin (Invivogen, ANTPM1). One million mouse embryonic stem cells (ESCs) carrying a previously published Nkx2-1mCherry reporter37,38 were first cultured in 10mL cSFDM in suspension for 60hr to allow embryonic body (EB) formation. Directed differentiation was performed by adapting our previously described approach for deriving airway organoids from mouse PSC.32,35,36 Briefly, to induce definitive endoderm fate, EBs were dissociated into a single cell suspension and cultured in cSFDM supplemented with 50ng/mL recombinant human/mouse/rat Activin A (R&D systems, 338-AC) for 60hr allowing EBs to reform in suspension culture. Next, 100ng/mL recombinant mouse Noggin (R&D systems, 1967-NG) and 10μM SB431542 (Sigma, S4317) were applied for 24hr to generate anterior foregut endoderm. Then, EBs were dissociated and single cell passaged onto a 6-well tissue culture plate coated with 200μL of growth factor reduced 3D-Matrigel (Corning, 354230). To induce NKX2-1 expression, adherent cells were cultured in cSFDM supplemented with 100ng/mL recombinant mouse Wnt3a (R&D systems, 1324-WN-010) and 10ng/mL recombinant human BMP4 (R&D systems, 314-BP) for 7-8 days depending on cell morphology and growth kinetics (typically 8 days; until total differentiation day D14). During the first 48hr (D6-8) the Wnt3a/BMP4 supplemented specification medium was further supplemented with 100nM Retinoic acid (Sigma, R2625) and 10μM Y-27632 (Tocris, 1254). On D13-14 the cells were incubated at 37°C for 1hr in 1mg/ml each of Collagenase IV (ThermoFisher, 17104019) and Dispase (Gibco, 17105-041) to digest the Matrigel bed, and the released epithelial spheres were isolated and washed with PBS through two slow centrifugations at 100xg. The resulting cell pellet was dissociated with 1mL warm 0.05% trypsin-EDTA (Gibco, 25300062) for 8min at 37°C, followed by inactivation with 1mL of cold fetal bovine serum (Gibco, 16141079). Cells were sorted by flow cytometry on the basis of EPCAM (BV421 anti-CD326, BD Biosciences, 563214) and Nkx2-1mCherry as previously described.35 EPCAM+/Nkx2-1mCherry+ cells were then resuspended in 3D-Matrigel at a density of 500 cells per μL and pipetted in 15-20 μL droplets onto the base of tissue culture plates.
After 20min at 37°C to allow gelling of the Matrigel, airway medium composed of cSFDM containing 250ng/mL recombinant human FGF2 (R&D systems, 233-FB), 100ng/mL recombinant human FGF10 (345-FG), 10μM Y27632, 100ng/mL Heparin (Sigma, H3393) to induce airway lineage. After 10-14 days, putative airway organoids were again dissociated into single cell suspension. The 3D-cultures were first incubated at 37°C in 1mg/mL dispase and 1mg/mL collagenase, then collected and centrifuged at 300xg for 5min. The cell pellet was then dissociated with 1mL warm 0.05% trypsin-EDTA for 8min at 37°C, and then inactivated with 1mL of cold fetal bovine serum. The cells were then strained through a 40μm cell filter and centrifuged at 300xg for 5min. Resulting cells were purified by flow cytometry sorting Nkx2-1mCherry+ cells and plated in 3D Matrigel for culturing in Basal Cell Medium (BCM) consisting of: a base medium of cSFDM supplemented with 250ng/mL recombinant human FGF2 (R&D systems, 233-FB), 100ng/mL recombinant human FGF10 (R&D systems, 345-FG), 10μM Y27632 (Tocris, 1254), 100ng/mL Heparin (Sigma, H3393), 1 μM A83-01 (Tocris, 2939) and 1 μM DMH-1 (Tocris, 4126). After 10-14 days, epithelial spheres outgrowths were single cell digested as before, and putative “iBCs” were purified by flow cytometry sorting Nkx2-1mCherry and NGFR (abcam, ab8875 or ab245134) double positive cells as indicated in the text. Nkx2-1mCherry+/NGFR+ cells were then re-plated in 3D Matrigel in BCM and serially passaged every 10-14 days without further sorting. Where indicated in the text, these cultures were either prepared for transplantation, characterization, or cryopreservation. For cryopreservation, cells were dissociated into single cells as described above and subsequently frozen in cryopreservation media (90%BCM + 10% DMSO) at 500,000 cells/mL at −150°C. To thaw cryopreserved frozen iBC archives, we warm the cryovial to 37°C in a water bath and dilute the cell suspension with 9mL of BCM. Cell suspensions were centrifuged at 300xg for 5 min, before resuspending in Matrigel at 500 cells per μL in 3D-Matrigel. After 10-15min of incubation at 37°C, BCM was added and iBC culture is maintained as described above.
Murine primary BC culture
Mouse tracheas (between larynx and carina) were harvested from adult mice (8-16 weeks). The lymphatic, muscular and other surrounding tissues were removed by surgical dissection, and the remaining tracheas were opened longitudinally. Tracheal epithelium was then dissociated by enzymatic digestion as previously described in Plasschaert et al.15 using 1.5mg/mL pronase (Roche, 10165921001) in Ham’s F12 at 4°C for 18hr to create a single cell suspension. To purify live tracheal epithelial cells, collected cells were stained with BV421-conjugated rat anti-mouse EPCAM monoclonal antibody (BV421 anti-CD326, BD Biosciences, 563214) and the cell viability dye DRAQ7 (Abcam, ab109202). DRAQ7-/EPCAMhi cells were sorted and resuspended in 3D-Matrigel at a density of 500 cells/μL and pipetted as 15-20μL droplets added onto the base of 12 well tissue culture plates. After 20 minutes of incubation at 37°C to allow gelling of the Matrigel, BCM was added with medium changes every two days of culture. Cells were passaged every 10-14 days with the same method used to passage murine iBC.
Establishing GFP or GFP-Luciferase tagged murine iBC lines via lentiviral transduction
Established iBCs (after the first NGFR sort) were used for lentiviral transduction as follows. Single cell suspensions were prepared as detailed above. Then, 100k cells were incubated in suspension with either pHAGE-EF1 αL-GFP-W lentivirus62 (Addgene #126686), or CMV-Luciferase-EF1α-copGFP lentivirus (System Biosciences, BLIV511PA-1) at a multiplicity of infection (MOI) of 50 in 200μL of BCM supplemented with 5μg/mL polybrene. Cells were incubated with lentiviral particles for 4hrs total, with the suspension gently agitated every 20min. Then, cells were diluted with 15mL of BCM, centrifuged at 300xg for 5min, and then washed with BCM again and pelleted for resuspension in 3D cultures as detailed above. The initial outgrowth after transduction was purified by flow sorting Nkx2-1mCherry+/NGFR+/GFP+ triple positive cells to establish each GFP or GFP-Luc tagged iBC line.
HBEC culture
HBECs (DD008P, DD024O) were received from the MLI Center Tissue Procurement and Cell Culture Core at the University of North Carolina (Director: Dr. Scott Randell). Human lung tissue was procured under the University of North Carolina Office of Research Ethics Biomedical Institutional Review Board Protocol No. 03-1396. Primary HBECs were cultured as described.63 Briefly, cryopreserved P1 HBECs were thawed and cultured on PureCol (Advanced Biomatrix, 5005)-coated tissue culture dish in BEGM media at two million cells in one 10cm plate. Two days after establishment of the culture, for GFP tagging, HBECs were transduced with EF1 αL-GFP-W lentivirus at MOI=10. Donor HBECs were cultured for a further 5-6 days in BEGM without passaging until the culture reached ~90% confluence, at which point HBECs were harvested for transplantation without further sorting.
Human iBC Culture
Human iBC lines (generated from previously characterized KOLF247 and BU3 NGPT29 iPSC lines) were derived by directed differentiation and cryopreserved or maintained as stable self-renewing cultures in serum-free, feeder-free medium as described previously.29 BU3 NGPT iPSCs were generated and maintained in house with Boston University Institutional Review Board approval #H33122. KOLF2-C1 clone D05 iPSC (46XY, normal karyotype by G-banding) were shared by Drs. Adam Williams and Justin McDonough (The Jackson Laboratory for Genomic Medicine).
Briefly, cryopreserved iBCs were thawed and cultured in 3D-Matrigel droplets in Pneumacult Ex-Plus media (StemCell Technologies, 5040) supplemented with A83-01, DMH-1, Y27632 and Primocin. Seven days after thawing, the 3D iBC cultures were dissociated in dispase and trypsin as described for mouse iBC cultures, and were transduced with pHAGE-EF1αL-tdTomato-W lentivirus at MOI of 10 for three hours in suspension, before replating in 3D-Matrigel at 400 cells/μL. After 10-14 days of culture, NGFR+/tdTomato+ were purified by flow cytometry and plated again in 3D-Matrigel at 400 cells/μL. After 14 days of further culture, the iBCs were harvested for transplantation without further cell sorting.
Reverse transcription quantitative PCR (RT-qPCR)
RNA was extracted by first lysing cells in QIAzol (Qiagen, 79306) or RLT Plus lysis buffer (QIAGEN, 1053393), and subsequently using the RNeasy Mini kit (Qiagen, 74104). Taqman Fast Universal PCR Master Mix (Applied Biosystems, 4364103) was used to reverse transcribe RNA into cDNA, followed by 40 cycles of real time quantitative PCR (qPCR) using Taqman probes (Table S6). Relative gene expression, normalized to 18S control, was calculated as fold change in 18S-normalized gene expression, over baseline, using the 2−ΔΔCT method.64 Baseline, defined as fold change = 1, was set to undifferentiated PSCs or primary control as indicated in the text. If undetected, a cycle number of 40 was assigned to allow fold change calculations.
Transplantation of BC/iBC culture
Adult recipient mice (8-16 weeks old) were used for all transplantation studies. Tracheal epithelial injury was induced in isoflurane anesthetized mice by intratracheal delivery of 20mL of 2% polidocanol using the tongue pull method, followed by recovery prior to cell delivery 5-5.5 hours later. Specifically, the recipient animal was anesthetized with 3% isoflurane until the animal developed exaggerated respiratory movements at rates of 1-1.5 breath/second. Then the mouse was lightly restrained by operator, while a second operator delivered polidocanol solution directly into the posterior oropharynx of the mouse. The nose was gently held to encourage the animal to aspirate through the mouse, about 2-4 times, or until the liquid was no longer visible. For cell transplants, mice were reanesthetized and 6x106 primary BCs or iBCs suspended in 20μL of DMEM were intratracheally instilled as above.
Mouse tissue harvest and whole mount fluorescence imaging
Recipient mice were sacrificed following isoflurane overdose and PBS perfusion through the right ventricle. Mouse tracheas were harvested from larynx to carina, and opened by longitudinal transection of the anterior trachea through the cartilaginous rings. For whole mount imaging, the opened trachea was placed on a glass slide and imaged by fluorescence microscopy using a Nikon Eclipse Ni-E microscope. Epifluorescence imaging quantification was performed with the Image J (Version 2.1.0/1.53i). Specifically, the “Color Threshold” function was used to select and calculate the area of GFP/RFP+ regions (based on GFP/RFP signal) as well as the entire recipient trachea (based on background autofluorescence). Transplantation efficiency was calculated by dividing the GFP/RFP+ area over total trachea area. To prepare 7 μm thick tissue sections for immunostaining, the tracheal tissue was then fixed with 4% PFA at 4C for 4 hours prior to embedding in paraffin.
Immunofluorescence microscopy
Fixed, paraffin-embedded mouse trachea or lung 7μm thick tissue sections were rehydrated using standard methods and treated with citric acid-based Antigen Unmasking Solution (Vector Laboratories, H-3300-250) according to the manufacturer’s instructions. Slides were washed in PBS and blocking with 10 % Normal donkey serum (Sigma, D9663), 2% Bovine Serum Albumin (Fisher Scientific, BP1600), and 0.5 % TritonX-100 (Sigma, T9284) for 1hr at room temperature. The M.O.M. blocking kit (Vector Laboratories, BMK-2202) was used when primary mouse antibodies were required. Sections were incubated in primary antibody (detailed in key resouces table) resuspended in blocking buffer diluted 1:4 in PBS overnight at 4°C, washed 3 times with PBS, and incubated in secondary antibody (detailed in key resouces table) resuspended in blocking buffer diluted 1:2 in PBS and Hoechst 33342 (1:1000, ThermoFisher) for 2hr at room temperature. Slides were subsequently washed 3 times with PBS and mounted using FluoroSave mounting reagent (Millipore, 345789). Slides were imaged using either Nikon Eclipse Ni-E microscope or Zeiss LSM710 confocal microscope.
Fluorescence-activated cell sorting (FACS) or analysis
Single cell suspensions were prepared as described for passaging (in vitro samples) or trachea digestion. When necessary, cells were stained with fluorescence-conjugated antibodies (detailed in key resouces table) for 30 minutes in FACS buffer (2% Fetal Bovine Serum in PBS) at 4°C and then resuspended in FACS buffer with 1:100 DRAQ7 or 1:1000 Calcein Blue, AM (ThermoFisher, C1429) (live/dead stain). FACS was performed on either a Beckman Coulter MoFLo Astrios or BD FACSARIA II SORP. Flow analysis was performed on a Beckman Coulter MoFLo Astrios or Stratedigm S1000EXi. Data analysis was performed with FlowJo software (version 10.8.1).
Serial secondary transplantation
GFP+ cells were harvested from transplant recipients 6–16 weeks post-transplantation, using the same method to harvest primary basal cells as described above. GFP+ cells were then isolated by FACS and cultured in BCM. To achieve the cell number necessary for the next round of transplantation, passage 0 outgrowth of GFP+ cells were passaged and purified on Nkx2-1mCherry+/GFP+/NGFR+ FACS gating after 10-14 days of culture in BCM. Then, the passage 1 outgrowth cells were transplanted into syngeneic recipients as described above.
Single cell RNA Sequencing
Single cell suspensions were prepared, and live epithelial cells were FACS purified on a Beckman Coulter MoFLo Astrios cell sorter as described above to collect populations described in the results section. Single-cell RNA-sequencing was performed using the Chromium Single Cell 3' system (10X Genomics) at the Single Cell Sequencing Core at Boston University Medical Center according to the manufacturer’s instructions (10X Genomics). Fastq files were generated using bcl2fastq v2.2 and Cellranger v.3.0.2. The sequenced files were mapped to the GRCm38/mm10 and/or GRCh30/hg38 supplemented with GFP, mCherry and tdTomato transcripts. We used Seurat v.3 to process the data further. Doublet rate was estimated according to the 10X chromium guidelines, in proportion to the density of cells loaded. These rates help to flag potential doublets based on their gene and UMO counts. Based on manufacturer’s recommendations for sequencing an average of >40,000 reads per cell to detect 1000-3000 genes per cell, sequencing depth was pursued to achieve these guidelines for all mouse cells, and dead cells or outliers were excluded by excluding any cells with fewer than 200 genes detected, as well as any cells with higher percentages of counts mapping to mitochondrial genes by setting a threshold of 15%, according to recommendation from the Seurat Package.65 For the human cell recipients, endogenous mouse cells were initially included in the sequencing experiment to generate cell numbers necessary for the technical processing of the sample. Therefore, these samples were mapped onto both GRCm38/mm10 and GRCh30/hg38. Post alignment, the Seurat object was created for both species. These objects were merged and then normalized using SCTransform, with cell degradation effect regressed out.66 After an initial linear dimensionality reduction (PCA), we used UMAP projections to represent the data and the Louvain algorithm was used for clustering without harmonization. Differential gene expression tests were done with MAST,67 with prior gene filters to reduce the burden of multiple test corrects (minimum gene detection percentage of 10% in at least one of the populations, minimum average log-fold change of 0.5 between the two populations). The data was then visualized using SPRING plot.44 In the case of in vivo samples, Louvain clustering was used to identify and remove non-epithelial cells. Samples from distinct runs were then combined without harmonization, and again clustered using Louvain clustering. For merged dataset analysis, Seurat objects of individual datasets were merged together without harmonization and analyzed.
In vivo bioluminescence imaging
The Xenogen IVIS Spectrum Instrument and Living Image Software Version 3.2 (Caliper LifeSciences) were used for imaging, analysis, and quantification of bioluminescence signals. The fur on the ventral side of the mice were removed by Nair prior to imaging. On the day of imaging, luciferin (Palmer Elmer, 122799, 15mg/kg body weight) was administered via intra-peritoneal injection. Animals were imaged immediately after the injection.
Cilia imaging
Mice were euthanized, perfused, and the trachea was harvested as described before. The apical luminal surfaces of the lung epithelia were labelled using a solution of PBS pH 7.4 supplemented with 1 μg/ml of Alexa Fluor 647 conjugated Wheat Germ Agglutinin (ThermoFisher, W32466) for 10 mins. This procedure was performed by insertion of a flexible cannula into the trachea which was held in place with a surgical suture, and the lungs were inflated using a reservoir held at 35cm above the surgical site. The trachea was removed by the release of the suture and further dissection above the branch point and butterflied across the tracheal rings and washed 3 times with PBS pH 7.4 before proceeding to imaging.
The labelled trachea was then placed with the luminal surface facing down between two glass slips held apart by glass spacers at 0.85mm in height. Motile cilia were imaged using a Zeiss LSM880 confocal microscope using the Fast Airyscan mode in line sequential mode using a Plan-Apochromat 20X/0.8 M27 objective at 1 frame per sec. Whole mounted imaging was performed by tilling in Z-stack mode using Airyscan mode in line sequential mode using a Plan-Apochromat 20X/0.8 M27 objective. The raw data was processed with ZEN Black. Time differential analysis and kymograph analysis was performed on acquired videos. Cilia length was calculated by measuring individual cilia while in orthogonal view of the 3D acquired images. Tissue sections were scanned at 20X using a ZEISS Axio Scan.Z1 slide scanner.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical methods relevant to RT-qPCR, immunostaining, or flow cytometry assessment are outlined in the figure legends. Paired, two-tailed Student’s t tests were used for comparisons involving only two groups, while one-way ANOVA was used when considering multiple groups. Statistical analysis was done in GraphPad Prism and p value<0.05 was used to determine statistical significance unless otherwise indicated in the text.
Supplementary Material
Highlights.
Directed differentiation of mouse PSCs generates airway basal stem cells (iBCs)
iBC transplantation reconstitutes the mouse airway basal stem cell compartment
Donor-derived cells maintain lifelong in vivo airway epithelial cell functions
Similar methods allow the engraftment of human primary or iPSC-derived basal cells
ACKNOWLEDGMENTS
We thank Brian R. Tilton and the entire BUSM Flow Cytometry Core and Dr. Yuriy Alekseyev and the entire BUSM Microarray and Sequencing Resource Core for their technical assistance. We thank the staff of the Marsico Lung Institute Tissue Procurement and Cell Culture Core for primary human cells and media. We would like to thank the entire Kotton Lab and Center for Regenerative Medicine for their support and suggestions throughout the course of this research. We are indebted to Greg Miller and Marianne James for overall laboratory support as well as for reprogramming and characterization of iPSC lines. We are also grateful to Dr. Laertis Ikonomou for his insight and input throughout the project. This work was supported by NHLBI Progenitor Cell Translational Consortium (PCTC) Jumpstart Award to L.M.; Boston University Kilachand Multi-cellular Design Program Accelerator Grants to L.M., M.J.H., and D.N.K; NIH grants U01HL134745, U01HL134766, U01HL148692, R01HL095993 to D.N.K.; NIH grant R01HL139799 and Cystic Fibrosis Foundation Grant HAWKIN20XX2 to F.J.H; NIH grant R01HL124392 and R21HD094012 to X.V.; NIH grant T32HL7035-44 and Cystic Fibrosis Foundation Grant to A.B.; NIH grant T32HL007035 to M.J.H; and NIH grant DK065988 and Cystic Fibrosis Grant BOUCHE19R0 to S.H.R.. Human cell biobanking and sharing was supported by NHLBI grant NO1: 75N92020C00005 to D.N.K. Graphical abstract was created with Biorender.com.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.stem.2023.07.014.
DECLARATION OF INTERESTS
The authors have filed a patent application related to the engraftment approach presented in this manuscript.
REFERENCES
- 1.Michalopoulos GK, and DeFrances MC (1997). Liver regeneration. Science 276, 60–66. 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 2.Barkauskas CE, Cronce MJ, Rackley CR, Bowie EJ, Keene DR, Stripp BR, Randell SH, Noble PW, and Hogan BLM (2013). Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest 123, 3025–3036. 10.1172/JCI68782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker N, Es J.H. van, Kuipers J, Kujala P, Born M. van den, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. (2007). Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007. 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 4.Dor Y, Brown J, Martinez OI, and Melton DA (2004). Adult pancreatic β-cells are formed by self-duplication rather than stem-cell differentiation. Nature 429, 41–46. 10.1038/nature02520. [DOI] [PubMed] [Google Scholar]
- 5.Blanpain C, Lowry WE, Geoghegan A, Polak L, and Fuchs E (2004). Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell 118, 635–648. 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, Randell SH, and Hogan BLM (2009). Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl. Acad. Sci. USA 106, 12771–12775. 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Connor N, Mulliken J, Banks-Schlegel S, Kehinde O, and Green H (1981). Grafting of burns with cultured epithelium prepared from autologous epidermal cells. Lancet 317, 75–78. 10.1016/S0140-6736(81)90006-4. [DOI] [PubMed] [Google Scholar]
- 8.Hirsch T, Rothoeft T, Teig N, Bauer JW, Pellegrini G, De Rosa LD, Scaglione D, Reichelt J, Klausegger A, Kneisz D, et al. (2017). Regeneration of the entire human epidermis using transgenic stem cells. Nature 551, 327–332. 10.1038/nature24487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rama P, Matuska S, Paganoni G, Spinelli A, De Luca MD, and Pellegrini G (2010). Limbal stem-cell therapy and long-term corneal regeneration. N. Engl. J. Med 363, 147–155. 10.1056/NEJMoa0905955. [DOI] [PubMed] [Google Scholar]
- 10.Sugimoto S, Kobayashi E, Fujii M, Ohta Y, Arai K, Matano M, Ishikawa K, Miyamoto K, Toshimitsu K, Takahashi S, et al. (2021). An organoid-based organ-repurposing approach to treat short bowel syndrome. Nature 592, 99–104. 10.1038/s41586-021-03247-2. [DOI] [PubMed] [Google Scholar]
- 11.Herriges MJ, Yampolskaya M, Thapa BR, Lindstrom-Vautrin J, Wang F, Na C-L, Ma L, Montminy MM, Huang J, Villacorta-Martin C, et al. (2023). Durable alveolar engraftment of PSC-derived lung epithelial cells into immunocompetent mice. Cell Stem Cell. 10.1016/j.stem.2023.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berical A, Lee RE, Randell SH, and Hawkins F (2019). Challenges facing airway epithelial cell-based therapy for cystic fibrosis. Front. Pharmacol 10, 74. 10.3389/fphar.2019.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rock JR, Randell SH, and Hogan BLM (2010). Airway basal stem cells: a perspective on their roles in epithelial homeostasis and remodeling. Dis. Model. Mech 3, 545–556. 10.1242/dmm.006031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montoro DT, Haber AL, Biton M, Vinarsky V, Lin B, Birket SE, Yuan F, Chen S, Leung HM, Villoria J, et al. (2018). A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 560, 319–324. 10.1038/s41586-018-0393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plasschaert LW, Žilionis R, Choo-Wing R, Savova V, Knehr J, Roma G, Klein AM, and Jaffe AB (2018). A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 560, 377–381. 10.1038/s41586-018-0394-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y, Riccio P, Schotsaert M, Mori M, Lu J, Lee DK, García-Sastre A, Xu J, and Cardoso WV (2018). Spatial-temporal lineage restrictions of embryonic p63+ progenitors establish distinct stem cell pools in adult airways. Dev. Cell 44, 752–761.e4. 10.1016/j.devcel.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans MJ, Winkle LSV, Fanucchi MV, and Plopper CG (2009). Cellular and molecular characteristics of basal cells in airway epithelium. Exp. Lung Res 27, 401–415. 10.1080/01902140120740. [DOI] [PubMed] [Google Scholar]
- 18.Hong KU, Reynolds SD, Watkins S, Fuchs E, and Stripp BR (2004). In vivo differentiation potential of tracheal basal cells: evidence for multipotent and unipotent subpopulations. Am. J. Physiol. Lung Cell Mol. Physiol 286. L643–L649. 10.1152/ajplung.00155.2003. [DOI] [PubMed] [Google Scholar]
- 19.Teixeira VH, Nadarajan P, Graham TA, Pipinikas CP, Brown JM, Falzon M, Nye E, Poulsom R, Lawrence D, Wright NA, et al. (2013). Stochastic homeostasis in human airway epithelium is achieved by neutral competition of basal cell progenitors. eLife 2, e00966. 10.7554/eLife.00966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watson JK, Rulands S, Wilkinson AC, Wuidart A, Ousset M, Van Keymeulen A, Göttgens B, Blanpain C, Simons BD, and Rawlins EL (2015). Clonal dynamics reveal two distinct populations of basal cells in slow-turnover airway epithelium. Cell Rep. 12, 90–101. 10.1016/j.celrep.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaughan AE, Brumwell AN, Xi Y, Gotts JE, Brownfield DG, Treutlein B, Tan K, Tan V, Liu FC, Looney MR, et al. (2015). Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature 517, 621–625. 10.1038/nature14112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nichane M, Javed A, Sivakamasundari V, Ganesan M, Ang LT, Kraus P, Lufkin T, Loh KM, and Lim B (2017). Isolation and 3D expansion of multipotent Sox9+ mouse lung progenitors. Nat. Methods 14, 1205–1212. 10.1038/nmeth.4498. [DOI] [PubMed] [Google Scholar]
- 23.Miller AJ, Hill DR, Nagy MS, Aoki Y, Dye BR, Chin AM, Huang S, Zhu F, White ES, Lama V, et al. (2018). In vitro induction and in vivo engraftment of lung bud tip progenitor cells derived from human pluripotent stem cells. Stem Cell Rep. 10, 101–119. 10.1016/j.stemcr.2017.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kathiriya JJ, Brumwell AN, Jackson JR, Tang X, and Chapman HA (2020). Distinct airway epithelial stem cells hide among club cells but mobilize to promote alveolar regeneration. Cell Stem Cell 26, 346–358.e4. 10.1016/j.stem.2019.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kathiriya JJ, Wang C, Zhou M, Brumwell A, Cassandras M, Le Saux CJL, Cohen M, Alysandratos KD, Wang B, Wolters P, et al. (2022). Human alveolar type 2 epithelium transdifferentiates into meta-plastic KRT5+ basal cells. Nat. Cell Biol 24, 10–23. 10.1038/s41556-021-00809-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liao CC, Chiu CJ, Yang YH, and Chiang BL (2022). Neonatal lung-derived SSEA-1+ cells exhibited distinct stem/progenitor characteristics and organoid developmental potential. Iscience 25, 104262. 10.1016/j.isci.2022.104262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Louie SM, Moye AL, Wong IG, Lu E, Shehaj A, Garcia-de-Alba C, Ararat E, Raby BA, Lu B, Paschini M, et al. (2022). Progenitor potential of lung epithelial organoid cells in a transplantation model. Cell Rep. 39, 110662. 10.1016/j.celrep.2022.110662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghosh M, Ahmad S, White CW, and Reynolds SD (2016). Transplantation of airway epithelial stem/progenitor cells: A future for cell-based therapy. Am. J. Respir. Cell Mol. Biol 56, 1–10. 10.1165/rcmb.2016-0181ma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hawkins FJ, Suzuki S, Beermann ML, Barillà C, Wang R, Villacorta-Martin C, Berical A, Jean JC, Le Suer JL, Matte T, et al. (2021). Derivation of airway basal stem cells from human pluripotent stem cells. Cell Stem Cell 28, 79–95.e8. 10.1016/j.stem.2020.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Longmire TA, Ikonomou L, Hawkins F, Christodoulou C, Cao Y, Jean JC, Kwok LW, Mou H, Rajagopal J, Shen SS, et al. (2012). Efficient derivation of purified lung and thyroid progenitors from embryonic stem cells. Cell Stem Cell 10, 398–411. 10.1016/j.stem.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCauley KB, Hawkins F, Serra M, Thomas DC, Jacob A, and Kotton DN (2017). Efficient Derivation of Functional Human Airway Epithelium from Pluripotent Stem Cells via Temporal Regulation of Wnt Signaling. Cell Stem Cell 20, 844–857.e6. 10.1016/j.stem.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCauley KB, Alysandratos KD, Jacob A, Hawkins F, Caballero IS, Vedaie M, Yang W, Slovik KJ, Morley M, Carraro G, et al. (2018). Single-cell transcriptomic profiling of pluripotent stem cell-derived SCGB3A2+ airway epithelium. Stem Cell Rep. 10, 1579–1595. 10.1016/j.stemcr.2018.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hawkins F, Kramer P, Jacob A, Driver I, Thomas DC, McCauley KB, Skvir N, Crane AM, Kurmann AA, Hollenberg AN, et al. (2017). Prospective isolation of NKX2-1–expressing human lung progenitors derived from pluripotent stem cells. J. Clin. Invest 127, 2277–2294. 10.1172/jci89950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacob A, Vedaie M, Roberts DA, Thomas DC, Villacorta-Martin C, Alysandratos K-D, Hawkins F, and Kotton DN (2019). Derivation of self-renewing lung alveolar epithelial type II cells from human pluripotent stem cells. Nat. Protoc 14, 3303–3332. 10.1038/s41596-019-0220-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ikonomou L, Herriges MJ, Lewandowski SL, Marsland R, Villacorta-Martin C, Caballero IS, Frank DB, Sanghrajka RM, Dame K, Kańduła MM, et al. (2020). The in vivo genetic program of murine primordial lung epithelial progenitors. Nat. Commun 11, 635. 10.1038/s41467-020-14348-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Serra M, Alysandratos KD, Hawkins F, McCauley KB, Jacob A, Choi J, Caballero IS, Vedaie M, Kurmann AA, Ikonomou L, et al. (2017). Pluripotent stem cell differentiation reveals distinct developmental pathways regulating lung-versus thyroid-lineage specification. Development 144, 3879–3893. 10.1242/dev.150193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bilodeau M, Shojaie S, Ackerley C, Post M, and Rossant J (2014). Identification of a proximal progenitor population from murine fetal lungs with clonogenic and multiline-age differentiation potential. Stem Cell Rep. 3, 634–649. 10.1016/j.stemcr.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurmann AA, Serra M, Hawkins F, Rankin SA, Mori M, Astapova I, Ullas S, Lin S, Bilodeau M, Rossant J, et al. (2015). Regeneration of thyroid function by transplantation of differentiated pluripotent stem cells. Cell Stem Cell 17, 527–542. 10.1016/j.stem.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mou H, Vinarsky V, Tata PR, Brazauskas K, Choi SH, Crooke AK, Zhang B, Solomon GM, Turner B, Bihler H, et al. (2016). Dual SMAD signaling inhibition enables long-term expansion of diverse epithelial basal cells. Cell Stem Cell 19, 217–231. 10.1016/j.stem.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller AJ, Yu Q, Czerwinski M, Tsai Y-H, Conway RF, Wu A, Holloway EM, Walker T, Glass IA, Treutlein B, et al. (2020). In vitro and in vivo development of the human airway at single-cell resolution. Dev. Cell 53, 117–128.e6. 10.1016/j.devcel.2020.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morrison SJ, Uchida N, and Weissman IL (1995). The biology of hematopoietic stem cells. Annu. Rev. Cell Dev. Biol 11, 35–71. 10.1146/annurev.cb.11.110195.000343. [DOI] [PubMed] [Google Scholar]
- 42.Borthwick DW, Shahbazian M, Krantz QT, Dorin JR, and Randell SH (2012). Evidence for stem-cell niches in the tracheal epithelium. Am. J. Respir. Cell Mol. Biol 24, 662–670. 10.1165/ajrcmb.24.6.4217. [DOI] [PubMed] [Google Scholar]
- 43.Engler AE, Ysasi AB, Pihl RMF, Villacorta-Martin C, Heston HM, Richardson HMK, Thapa BR, Moniz NR, Belkina AC, Mazzilli SA, et al. (2020). Airway-associated macrophages in homeostasis and repair. Cell Rep. 33, 108553. 10.1016/j.celrep.2020.108553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinreb C, Wolock S, and Klein AM (2018). SPRING: a kinetic interface for visualizing high dimensional single-cell expression data. Bioinformatics 34, 1246–1248. 10.1093/bioinformatics/btx792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guha A, Vasconcelos M, Zhao R, Gower AC, Rajagopal J, and Cardoso WV (2014). Analysis of Notch signaling-dependent gene expression in developing airways reveals diversity of Clara cells. PLoS ONE 9, e88848. 10.1371/journal.pone.0088848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siminovitch L, Till JE, and McCulloch EA (1964). Decline in colony-forming ability of marrow cells subjected to serial transplantation into irradiated mice. J. Cell. Comp. Physiol 64, 23–31. 10.1002/jcp.1030640104. [DOI] [PubMed] [Google Scholar]
- 47.Skarnes WC, Pellegrino E, and McDonough JA (2019). Improving homology-directed repair efficiency in human stem cells. Methods 164–165, 18–28. 10.1016/j.ymeth.2019.06.016. [DOI] [PubMed] [Google Scholar]
- 48.Pollock K, Albares L, Wendt C, and Hubel A (2013). Isolation of fibroblasts and epithelial cells in bronchoalveolar lavage (BAL). Exp. Lung Res 39, 146–154. 10.3109/01902148.2013.781720. [DOI] [PubMed] [Google Scholar]
- 49.Butler CR, Hynds RE, Gowers KHC, Lee DDH, Brown JM, Crowley C, Teixeira VH, Smith CM, Urbani L, Hamilton NJ, et al. (2016). Rapid expansion of human epithelial stem cells suitable for airway tissue engineering. Am. J. Respir. Crit. Care Med 194, 156–168. 10.1164/rccm.201507-1414oc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gentzsch M, Boyles SE, Cheluvaraju C, Chaudhry IG, Quinney NL, Cho C, Dang H, Liu X, Schlegel R, and Randell SH (2016). Pharmacological rescue of conditionally reprogrammed cystic fibrosis bronchial epithelial cells. Am. J. Respir. Cell Mol. Biol 56, 568–574. 10.1165/rcmb.2016-0276ma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang C, Lee HJ, Shrivastava A, Wang R, McQuiston TJ, Challberg SS, Pollok BA, and Wang T (2018). Long-term in vitro expansion of epithelial stem cells enabled by pharmacological inhibition of PAK1-ROCK-myosin II and TGF-β signaling. Cell Rep. 25, 598–610.e5. 10.1016/j.celrep.2018.09.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crystal RG (2014). Airway basal cells. The “smoking gun” of chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med 190, 1355–1362. 10.1164/rccm.201408-1492PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crane AM, Kramer P, Bui JH, Chung WJ, Li XS, Gonzalez-Garay ML, Hawkins F, Liao W, Mora D, Choi S, et al. (2015). Targeted correction and restored function of the CFTR gene in cystic fibrosis induced pluripotent stem cells. Stem Cell Rep. 4, 569–577. 10.1016/j.stemcr.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abo KM, Sainz de Aja JS, de Lindstrom-Vautrin J, Alysandratos KD, Richards A, Garcia-de-Alba C, Huang J, Hix OT, Werder RB, Bullitt E, et al. (2022). Air-liquid interface culture promotes maturation and allows environmental exposure of pluripotent stem cell–derived alveolar epithelium. JCI Insight 7, e155589. 10.1172/jci.insight.155589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alysandratos K-D, Rivas C.G.de A., Yao C, Pessina P, Villacorta-Martin C, Huang J, Hix OT, Minakin K, Konda B, Stripp BR, et al. (2022). Impact of cell culture on the transcriptomic programs of primary and iPSC-derived human alveolar type 2 cells. JCI Insight 8, e158937. 10.1101/2022.02.08.479591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pavlovic BJ, Blake LE, Roux J, Chavarria C, and Gilad Y (2018). A comparative assessment of human and chimpanzee iPSC-derived cardiomyocytes with primary heart tissues. Sci. Rep 8, 15312. 10.1038/s41598-018-33478-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwartz RE, Fleming HE, Khetani SR, and Bhatia SN (2014). Pluripotent stem cell-derived hepatocyte-like cells. Biotechnol. Adv 32, 504–513. 10.1016/j.biotechadv.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xia N, Zhang P, Fang F, Wang Z, Rothstein M, Angulo B, Chiang R, Taylor J, and Reijo Pera RAR (2016). Transcriptional comparison of human induced and primary midbrain dopaminergic neurons. Sci. Rep 6, 20270. 10.1038/srep20270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fidanza A, Stumpf PS, Ramachandran P, Tamagno S, Babtie A, Lopez-Yrigoyen M, Taylor AH, Easterbrook J, Henderson BEP, Axton R, et al. (2020). Single-cell analyses and machine learning define hematopoietic progenitor and HSC-like cells derived from human PSCs. Blood 136, 2893–2904. 10.1182/blood.2020006229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fujimura T, Enomoto Y, Katsura H, Ogawa T, Baba S, Ogata A, Yamaoka A, Shiroguchi K, and Morimoto M (2023). Identifying a lung stem cell subpopulation by combining single-cell morphometrics, organoid culture, and transcriptomics. Stem Cellssxad044. 10.1093/stmcls/sxad044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dorrello NV, Guenthart BA, O’Neill JD, Kim J, Cunningham K, Chen YW, Biscotti M, Swayne T, Wobma HM, Huang SXL, et al. (2017). Functional vascularized lung grafts for lung bioengineering. Sci. Adv 3, e1700521. 10.1126/sciadv.1700521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilson AA, Murphy GJ, Hamakawa H, Kwok LW, Srinivasan S, Hovav AH, Mulligan RC, Amar S, Suki B, and Kotton DN (2010). Amelioration of emphysema in mice through lentiviral transduction of long-lived pulmonary alveolar macrophages. J. Clin. Invest 120, 379–389. 10.1172/JCI36666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fulcher ML, and Randell SH (2012). Human nasal and tracheo-bronchial respiratory epithelial cell culture. Methods Mol. Biol 945, 109–121. 10.1007/978-1-62703-125-7_8. [DOI] [PubMed] [Google Scholar]
- 64.Pfaffl MW (2001). A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29, e45. 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. (2021). Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29. 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, et al. (2015). MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278. 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. The sequencing data will also be available on the Kotton Lab’s Bioinformatics Portal at https://www.kottonlab.com. This paper does not report original code. Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit Anti-NGFR Antibody (no longer in production) | Abcam | Cat#ab8875; RRID:AB_306828 |
| Mouse Anti-NGFR Antibody | Abcam | Cat#ab245134 |
| BV421 Rat Anti-Mouse CD326 | BD Biosciences | Cat#563214; RRID:AB_2738073 |
| Goat Anti-GFP Antibody | US Biological | Cat#G8965-01E |
| Chicken Anti-GFP Antibody | Aves Labs | Cat#GFP-1020; RRID:AB_10000240 |
| Goat Anti-RFP Antibody | My Biosource | Cat#MBS448122 |
| Rat Anti-Mki67 Antibody | Thermo Fisher | Cat#14-5698-82; RRID:AB_10854564 |
| Rabbit Anti-NKX2-1 Antibody | Abcam | Cat#ab76013; RRID:AB_1310784 |
| Rabbit Anti-KRT5 Antibody | BioLegend | Cat#905501; RRID:AB_2565050 |
| Rabbit Anti-RFP Antibody | Rockland | Cat#600-401-379; RRID:AB_2209751 |
| Mouse Anti-Acetylated-Tubulin Antibody | Sigma | Cat#T7451; RRID:AB_609894 |
| Goat Anti-SCGB1A1 (mouse) Antibody | Gift from Barry Stripp | N/A |
| Goat Anti-SCGB1A1 (human) Antibody | Sigma | Cat#ABS1673; RRID:AB_2910611 |
| Rat Anti-E-Cadherin Antibody | Invitrogen | Cat#13-1900; RRID:AB_86571 |
| Alexa Fluor 488 donkey anti-chicken IgY (IgG) (H+L) | Jackson Immuno Research | Cat#703-545-155; RRID:AB_2340375 |
| Alexa Fluor 488 donkey anti-goat IgG (H+L) | Jackson Immuno Research | Cat#705-545-003; RRID:AB_2340428 |
| Alexa Fluor 488 donkey anti-rat IgG (H+L) | Jackson Immuno Research | Cat#712-546-153; RRID:AB_2340686 |
| Alexa Fluor 488 donkey anti-rabbit IgG (H+L) | Jackson Immuno Research | Cat#711-546-152; RRID:AB_2340619 |
| Alexa Fluor 488 donkey anti-mouse IgG (H+L) | Jackson Immuno Research | Cat#715-545-150; RRID:AB_2340846 |
| Alexa Fluor 546 donkey anti-Rabbit IgG (H+L) | Thermo Fisher | Cat#A11055; RRID:AB_2534102 |
| Alexa Fluor 568 donkey anti-mouse IgG (H+L) | Thermo Fisher | Cat#A10037; RRID:AB_2534013 |
| Cy3 donkey anti-rat IgG (H+L) | Jackson Immuno Research | Cat#712-166-153; RRID:AB_2340669 |
| Cy3 donkey anti-rabbit IgG (H+L) | Jackson Immuno Research | Cat#711-165-152; RRID:AB_2307443 |
| Cy3 donkey anti-goat IgG (H+L) | Jackson Immuno Research | Cat#705-165-147; RRID:AB_2307351 |
| Alexa Fluor 647 donkey anti-rabbit IgG (H+L) | Thermo Fisher | Cat#A32795; RRID:AB_2762835 |
| Alexa Fluor 647 donkey anti-goat IgG (H+L) | Jackson Immuno Research | Cat#705-605-003; RRID:AB_2340436 |
| Bacterial and virus strains | ||
| pHAGE-EF1αL-GFP-W lentivirus | Darrell Kotton Lab | Addgene126686 |
| pHAGE-EF1αL-tdTomato-W lentivirus | Darrell Kotton Lab | N/A |
| CMV-Luciferase-EF1α-copGFP lentivirus | System Biosciences | Cat#BLIV511PA-1 |
| Biological samples | ||
| Primary human airway epithelial cell culture | The MLI Tissue Procurement and Cell Culture Core, University of North Carolina, Chapel Hill | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| DMEM | Gibco | Cat#2414671 |
| IMDM | ThermoFisher | Cat#12440053 |
| Glutamax | Life Technologies | Cat#35050-061 |
| Ham’s F12 | Cellgro | Cat#10-080-CV |
| B27 supplement | Invitrogen | Cat#17504-44 |
| N2 Supplement | Invitrogen | Cat#17502-048 |
| Primocin | Invivogen | Cat#ANTPM1 |
| 7.5% BSA Fraction V | ThermoFisher | Cat#15260-037 |
| 1-thioglycerol (MTG) | Sigma | Cat#M6145 |
| Ascorbic Acid | Sigma | Cat#A4544 |
| Recombinant human/mouse/rat Activin A | R&D Systems | Cat#338-AC |
| Recombinant mouse Wnt3a | R&D Systems | Cat#1324-WN-010 |
| Recombinant human Bmp4 | R&D Systems | Cat#314-BP |
| Recombinant mouse Noggin | R&D Systems | Cat#1967-NG |
| SB431542 | Sigma | Cat#S4317 |
| Recombinant human FGF2 | R&D Systems | Cat#233-FB |
| Recombinant human FGF10 | R&D Systems | Cat#345-FG |
| DMH-1 | Tocris | Cat#4126 |
| A8301 | Tocris | Cat#2939 |
| Y-27632 (ROCK inhibitor) | Tocris | Cat#1254 |
| Heparin Salt | Sigma Millipore | Cat#H3393 |
| Growth Factor Reduced 3D matrigel | Corning | Cat#354230 |
| Dispase | Gibco | Cat#17105-041 |
| Collagenase Type IV | ThermoFisher | Cat#17104019 |
| Pneumacault ExPlus Medium | Stem Cell Technologies | Cat#5040 |
| BEGM | The MLI Tissue Procurement and Cell Culture Core, University of North Carolina, Chapel Hill | N/A |
| Retinoic acid | Sigma | Cat#R2625 |
| 0.05% Trypsin-EDTA | Gibco | Cat#25300062 |
| fetal bovine serum | Gibco | Cat#16141079 |
| PureCol | Advanced Biomatrix | Cat#5005 |
| Paraformaldehyde | Electron Microscopy Sciences | Cat#19208 |
| Normal Donkey Serum | Sigma | Cat#D9663 |
| Antigen Unmasking Solution, Citric Acid Based | Vector Laboratories | Cat#H-3300-250 |
| M.O.M. Immunodetection Kit | Vector Laboratories | Cat#BMK-2202 |
| Bovine Serum Albumin | Fisher | Cat#BP1600 |
| FluoroSave Mounting Agent | Millipore | Cat#345789 |
| Hoecsht 33342 | Thermo Fisher | Cat#H3570 |
| Triton-X | Sigma | Cat#T9284 |
| Pronase | Roche | Cat#10165921001 |
| Wheat Germ Agglutinin, Alexa Fluor™ 647 Conjugate | ThermoFisher | Cat#W32466 |
| DNAse | Roche | Cat#10104159001 |
| Polidocanol | Sigma | Cat#88315 |
| XenoLight D-Luciferin | PerkinElmer | Cat#122799 |
| Calcein Blue | ThermoFisher | Cat#C1429 |
| DRAQ7 | Abcam | Cat#ab109202 |
| Critical commercial assays | ||
| RNeasy Mini Kit | QIAGEN | Cat#74104 |
| QIAzol Lysis Reagent QIAGEN | QIAGEN | Cat#79306 |
| RLT Plus lysis buffer | QIAGEN | Cat#1053393 |
| TaqMan Fast Universal PCR Master Mix (2X) | Thermo Fisher | Cat#4364103 |
| High-Capacity cDNA Reverse Transcription Kit | Applied Biosystems | Cat#4368814 |
| Click-iT EdU Cell Proliferation Kit | ThermoFisher | Cat#C10340 |
| Deposited data | ||
| Sequence Data | This Paper | GEO: Super Series GSE211998 |
| Experimental models: Cell lines | ||
| Nkx2-1mCherry mouse ES Cell Line | Bilodeau et al.37 | N/A |
| Human BU3-NGPT iPSC Cell Line | Hawkins et al.29 | N/A |
| Human KOLF2 iPSC Cell Line | Skarnes et al.47 | N/A |
| Experimental models: Organisms/strains | ||
| UBC-GFP mice | Jackson Labs | JAX004353 |
| C57BL/6J mice | Jackson Labs | JAX000664 |
| 129X1/SvJ mice | Jackson Labs | JAX000691 |
| 129S1/SvlmJ mice | Jackson Labs | JAX002448 |
| NSG mice | Jackson Labs | JAX005557 |
| Software and algorithms | ||
| FlowJo Software v.10.8.1 | Becton Dickinson & Company | https://www.flowjo.com/solutions/flowjo |
| Seurat v.3 | Sajita Lab65 | https://github.com/satijalab/seurat |
| SRPING | Klein Lab44 | https://github.com/AllonKleinLab/SPRING_dev |
| Living Image Software v. 3.2 | Caliper LifeSciences | https://www.perkinelmer.com/product/spectrum-200-living-image-v4series-1-128113 |
| Graphpad Prism v.9.5.1 | GraphPad Software | https://www.graphpad.com/features |
| ImageJ v.2.1.0/1.53i | NIH | https://imagej.net/ij/index.html |
| Other | ||
| Taqman Probe: Nkx2-1 | Thermo Fisher | Mm00447558_m1 |
| Taqman Probe: Trp63 | Thermo Fisher | Mm00495788_m1 |
| Taqman Probe: Krt5 | Thermo Fisher | Mm01305291_g1 |
| Taqman Probe: Ngfr | Thermo Fisher | Mm00446296_m1 |
| Taqman Probe: Scgb1a1 | Thermo Fisher | Mm00442046_m1 |
| Taqman Probe: Muc5ac | Thermo Fisher | Mm01276718_m1 |
| Taqman Probe: Foxj1 | Thermo Fisher | Mm01267279_m1 |