Fig. 1. Transcriptional regulation by influenza-programmed Cas9 (TRPPC) manipulates host gene expression to enable fitness-based screening.
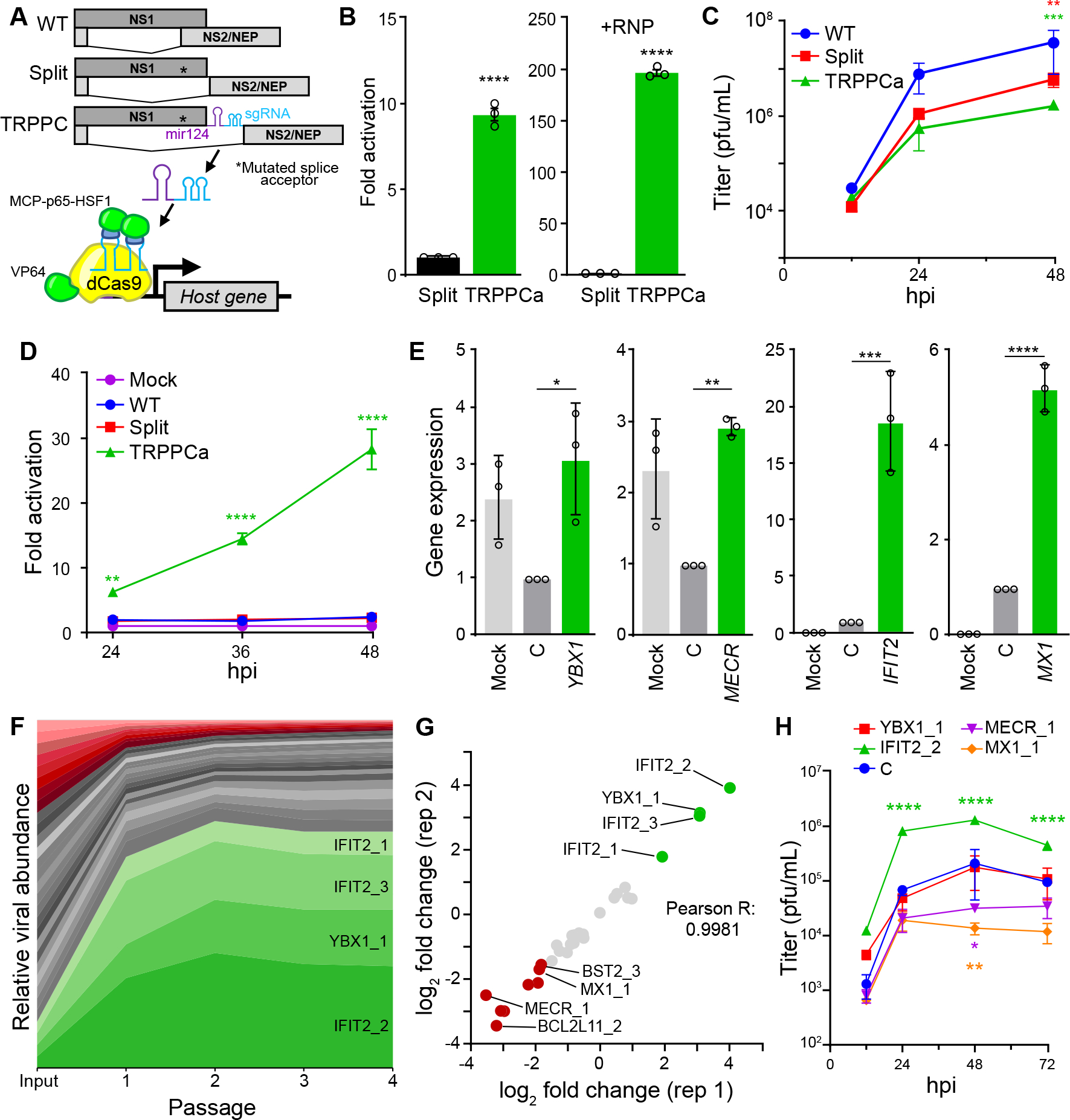
A, Cartoon of re-engineered TRPPC NS genome segment and TRPPCa-mediated gene expression. The sgRNA directs VP64-dCas9 to specific genome targets while two MS2 hairpins inserted in the sgRNA recruit MCP-p65-HSF1.
B, TRPPCa of a luciferase reporter in 293T cells. Cell were transfected with vectors expressing viral genomic RNA for NS, Split NS that lacks an sgRNA, or TRPCC NS targeting the reporter promoter. Activation was measured in the presence (+RNP, right) or absence (left) of the viral replication machinery.
C, Multicycle replication of IAV harboring a TRPPC-NS segment in A549 cells.
D, Virally delivered sgRNA activates reporter gene expression in a multicycle infection. A549-CRISPRa cells were inoculated with virus encoding the indicated NS segment (MOI = 0.05), or mock treated, and luciferase reporter was measured over the course of infection.
E, Virally delivered sgRNAs activate expression of host genes from the endogenous locus. A549-CRISPRa cells were inoculated with TRPPC viruses (MOI = 5) targeting the indicated gene, a non-targeting control (C) or mock. Host gene expression was measured at 8 hpi via RT-qPCR.
F, A pool of 34 TRPPC viruses targeting a collection of 11 potential pro- or antiviral host genes were subject to 4 rounds of selection in A549-CRISPRa cells cells. Viruses present at each stage of selection were quantified by deep-sequencing and normalized sgRNA composition is depicted. Viruses activating proviral genes enriched at least 3-fold are colored green, while viruses activating antiviral genes that are depleted at least 3-fold are colored red. Graph is representative of mean values for 2 replicate screens.
G, TRPPCa screens are highly reproducible. Comparison of two biological replications shows nearly identical relative enrichment of TRPPC viruses targeting the indicated host genes after 4 rounds of selection.
H, TRPPC results reflect changes in viral replication. Multicycle replication in A549-CRISPRa cells of individual TRPPC viruses targeting specific host genes (MOI = 0.01).
Data are shown as grand mean of 3 replicates ± SEM (B, D) or mean ± s.d. (C, E, F, H). T tests (B), two-way ANOVA with Dunnett’s multiple comparisons test against WT (C, D, H), and one-way ANOVA with Dunnett’s multiple comparisons tests (E) were performed (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001).
See also Figure S1.
