Figure 3. Mendelian randomization (MR) analyses showing genetic associations between leukocyte telomere length and age at natural menopause.
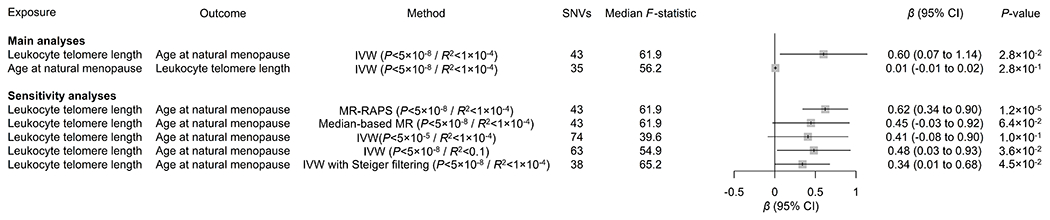
Main MR analyses used the inverse-variance-weighted (IVW) method, using genome-wide significant (P<5×10−8) single-nucleotide variants (SNVs) clumped into independent loci using a linkage disequilibrium R2 threshold of 1×10−4. Sensitivity analyses used the MR with robust adjusted profile score (MR-RAPS) and median-based MR methods, as well as the IVW method using more lenient P-value (5.0×10−5) or linkage disequilibrium R² (1.0×10−1) thresholds. Genetic data were expressed per standard deviation of (increased) leukocyte telomere length and per year of (older) age at natural menopause. No corrections for multiple comparisons were made. CI indicates confidence interval.
