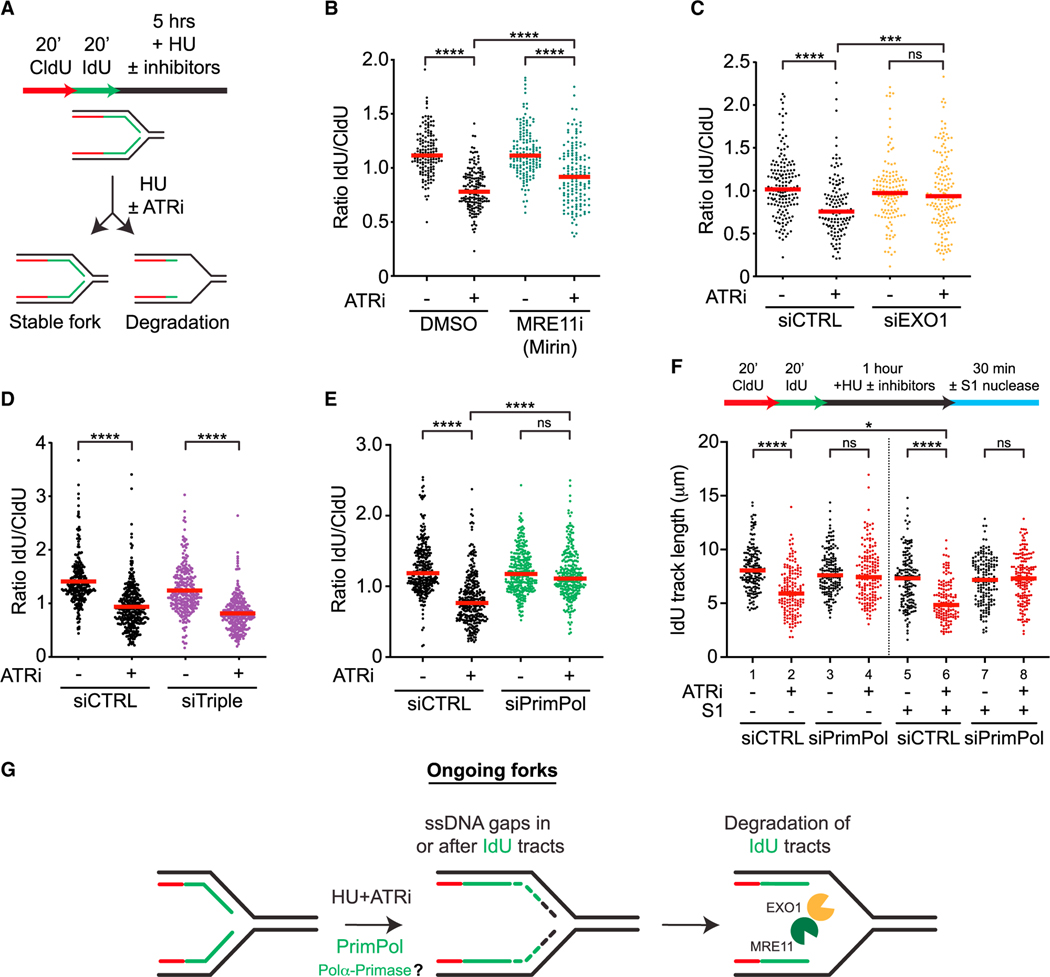
Figure 2. ATR prevents resection from PrimPol-generated gaps at ongoing forks.
(A) Schematic representation of the nascent DNA degradation fiber assay.
(B and C) Cells were treated as in (A) with or without ATRi (VE-821, 10 μM) and MRE11i (Mirin, 50 μM) in (B), and with or without ATRi (VE-821, 10 μM) following 48 h siRNA knockdown of EXO1 in (C). Number (n) of fibers quantified >250 across two biological replicates. Significance was calculated using the Mann-Whitney Ranked Sum Test with ***p < 0.001, ****p < 0.0001.
(D and E) siRNAs against HLTF, SMARCAL1, and ZRANB3 (siTriple) (D) or PrimPol (E) were transfected 48 h prior to fiber analysis. Number (n) of fibers quantified >500 across three biological replicates. Significance was calculated using the Mann-Whitney Ranked Sum Test with ****p < 0.0001.
(F) U2OS cells were analyzed as in (E) except cells were treated for only 1 h with or without ATRi (VE-821, 10 μM) and HU (4 mM) followed by S1 nuclease digestion. Number (n) of fibers quantified >135 in each sample across two biological replicates. Significance was calculated using the Mann-Whitney Ranked Sum Test with *p < 0.05, ****p < 0.0001.
(G) Model for how nascent DNA is degraded at ongoing forks.