Figure 2: ESC-derived Tip-like Cells are Morphologically and Transcriptionally Similar to Cultured Primary Tip-like Cells.
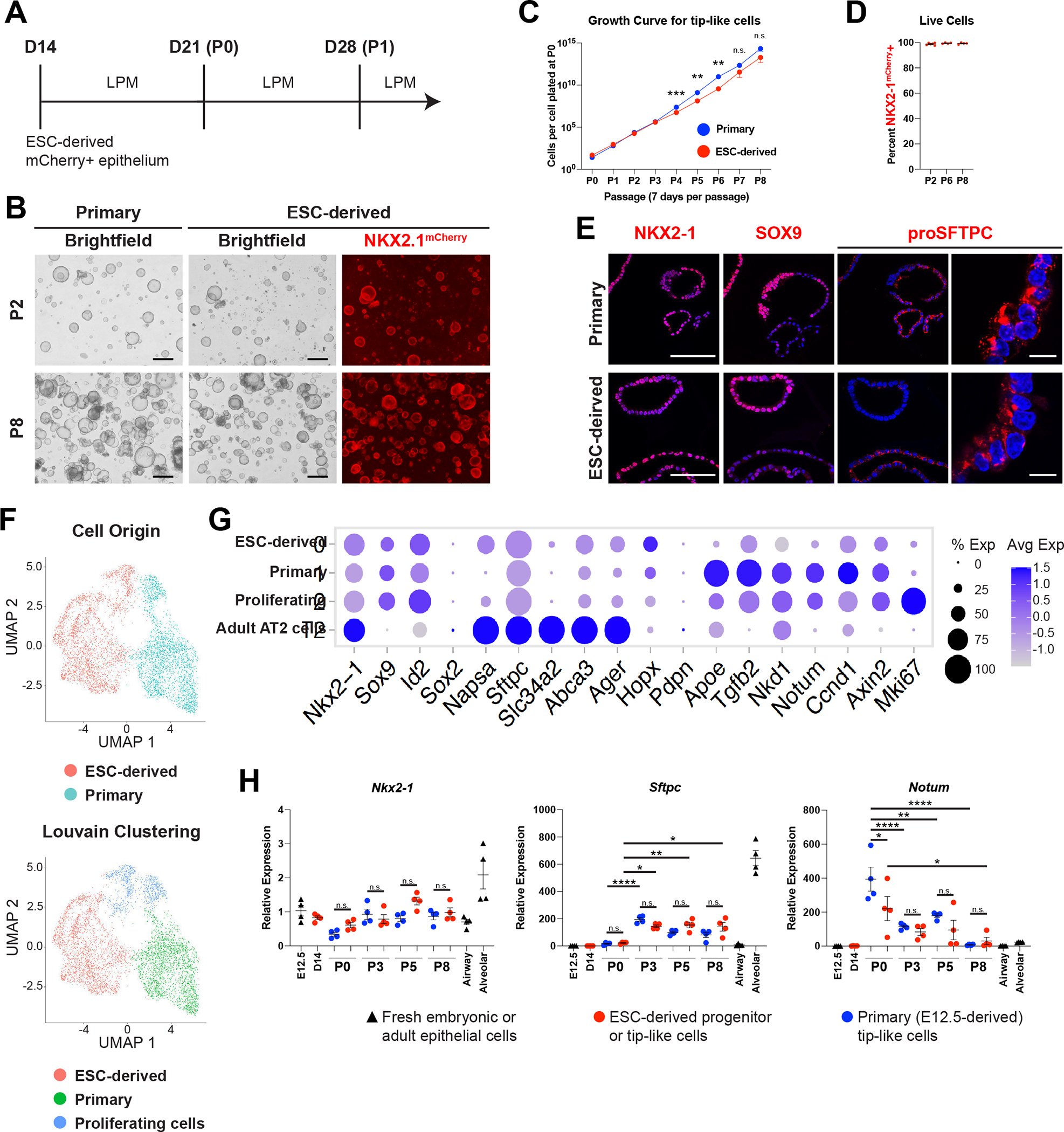
(A) Schematic of differentiation and passaging of ESC-derived tip-like cells in lung progenitor medium (LPM) following cell sorting of Nkx2–1mCherry+ cells on day 14.
(B) Fluorescent microcopy images of primary and ESC-derived tip-like cells at P2 and P8. Scale bars are 500um.
(C) Quantification of cell proliferation for primary and ESC-derived tip-like cells across nine passages. n.s. not significant, ** p<0.01, *** p<0.001 by unpaired, two-tailed Student’s t-test. n= 4 biological replicates. Error bars= mean +/− SEM
(D) Assessment of NKX2–1mCherry expression throughout passaging of ESC-derived tip-like cells. n= 4 biological replicates. Error bars = mean +/− SEM.
(E) Immunofluorescence microscopy for NKX2–1, SOX9, and proSFTPC in primary and ESC-derived tip-like cells. Nuclei stained with Hoechst, scale bars are 100um (first three columns) or 10um (rightmost column).
(F) UMAP plot for scRNA-seq of primary and ESC-derived tip-like cells. Top plot distinguishes cells by sample origin; bottom plot displays Louvain clusters.
(G) Expression of genes, including those identified as differently expressed between primary and ESC-derived tip-like cells. Cells from clusters annotated in panel F are compared against primary adult AT2 cells collected and sequenced at the same time.
(H) Analysis of gene expression by RT-qPCR. Primary and ESC-derived tip-like cells from multiple passages are compared against lung epithelial progenitors from day 14 of the WBRF protocol (D14) and freshly sorted lung epithelial cells from embryonic (E12.5) and adult (Airway and Alveolar) mouse lungs. Reference gating for primary controls can be found in supplemental figure 2C,D. n.s. not significant, * p<0.05, ** p<0.01, **** p<0.0001 by one-way ANOVA. n= 4 biological replicates. Error bars = mean +/− SEM.
See also Figures S1, S2 and Tables S1, S2.
