SUMMARY
Nutrient starvation drives yeast meiosis, whereas retinoic acid (RA) is required for mammalian meiosis through its germline target Stra8. Here, by using single-cell transcriptomic analysis of wild-type and Stra8-deficient juvenile mouse germ cells, our data show that the expression of nutrient transporter genes, including Slc7a5, Slc38a2, and Slc2a1, is downregulated in germ cells during meiotic initiation, and this process requires Stra8, which binds to these genes and induces their H3K27 deacetylation. Consequently, Stra8-deficient germ cells sustain glutamine and glucose uptake in response to RA and exhibit hyperactive mTORC1/protein kinase A (PKA) activities. Importantly, expression of Slc38a2, a glutamine importer, is negatively correlated with meiotic genes in the GTEx dataset, and Slc38a2 knockdown downregulates mTORC1/PKA activities and induces meiotic gene expression. Thus, our study indicates that RA via Stra8, a chordate morphogen pathway, induces meiosis partially by generating a conserved nutrient restriction signal in mammalian germ cells by downregulating their nutrient transporter expression.
Graphical abstract
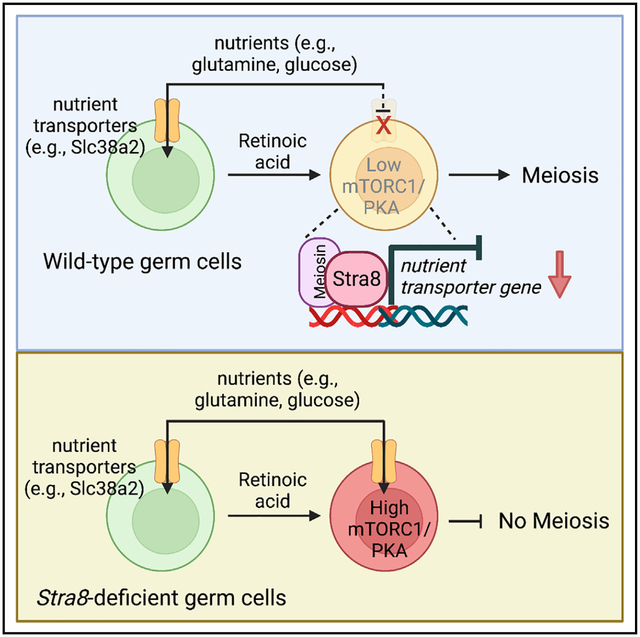
In brief
Nutrient restriction induces meiosis in yeast. Here, Zhang et al. show that chordate morphogen retinoic acid via its germline target Stra8 induces downregulation of nutrient transporter expression during meiotic initiation in male mouse germ cells, thereby eliciting a conserved nutrient restriction signal including mTORC1/PKA inactivation to activate meiotic gene expression.
INTRODUCTION
The foundation of sexual reproduction is haploid gametes. To form haploid gametes, germline stem cells must cease mitosis and transition into meiosis, a specialized cell cycle that has been widely conserved throughout evolution.1 During meiosis, one round of DNA replication is followed by two rounds of chromosome segregation. Additionally, homologous recombination that takes place during prophase in meiosis I involves the exchange of genetic materials between maternal and paternal alleles, resulting in new combinations of genes.
In Saccharomyces cerevisiae, the decision to enter meiosis is controlled by nutrient availability and mating-type signals, which regulate the expression of the master transcription factor for meiotic entry, IME1.2–4 When nutrients are available, several transcription factors mediate the association of the transcription repressor Tup1 with the IME1 promoter. When nutrients become limited, these transcription factors and Tup1 dissociate, resulting in active IME1 transcription and meiotic initiation.
In mammals, while female meiosis takes place in developing ovaries, male meiosis occurs in male germ cells that reside inside the seminiferous tubules of postnatal testes upon puberty. Undifferentiated spermatogonia reside near the basement membrane and consist of single cells (As spermatogonia, often referred to as spermatogonial stem cells [SSCs]) and chains of 2–16 linked spermatogonia (termed Apr and Aal,).5,6 Undifferentiated spermatogonia differentiate to form A1 spermatogonia, which further undergo five additional rounds of mitotic division to form B spermatogonia. The B spermatogonia then divide and differentiate into preleptotene spermatocytes that enter meiosis as leptotene spermatocytes. Interestingly, the transition from mitosis to meiosis is also associated with the migration of male germ cells across an ultrastructure—the blood-testis barrier (BTB)—that is formed by the tight junctions between Sertoli cells inside the seminiferous tubule.7 Retinoic acid (RA), a chordate morphogen, is required for the onset of mammalian meiosis in both sexes through its germline target, stimulated by retinoic acid gene 8 (Stra8).8–10 Mechanistically, Stra8 has been shown to bind to the regulatory sequences of thousands of genes and is shown to cooperate with another germline-specific cofactor, Meiosin.11,12
We have reported that an autophagy-inducing factor acts on meiosis-initiating germ cells in both sexes.13 Interestingly, nutrient restriction, an inducer of yeast meiosis, is perhaps the most potent inducer of autophagy.14 Indeed, under nutrient-restricted conditions, our past work15 has shown that RA induces meiosis in cultured male germline or SSCs in vitro, suggesting that nutrient restriction is a conserved inducer of meiosis. These studies raise the following interesting questions: while yeasts passively depend on environmental nutrient availability for their meiotic decision, how do mammalian germ cells “starve” themselves? In this study, we unexpectedly found that mammalian germ cells downregulate the expression of their nutrient transporters during meiotic initiation to limit nutrient uptake, and this process is mediated by RA via Stra8. Our results suggest a cell-autonomous mechanism of programmed “starvation” in mammalian germ cells for regulated meiotic initiation.
RESULTS
scRNA-seq analysis of wild-type and Stra8-deficient juvenile testes
We collected mouse testicular cells from wild-type and Stra8-deficient mice at postnatal day 12 (P12) and P21 for single-cell RNA sequencing (scRNA-seq). Upon scRNA-seq, we obtained a total of 23,470 cells from these samples. Using stringent quality control (Figure S1), 22,839 cells were selected for analysis. All cells, including both wild-type and Stra8-deficient testicular cells at P12 and P21, were pooled together to perform clustering analysis. The results revealed six major cell clusters based on their distinct gene-expression patterns (Figure S2A). We annotated these six clusters by using known marker genes (Figures S2B and S2C), including spermatogonia and early spermatocytes (SPGC; 3,941 cells), spermatocytes (SPC; 4,924 cells), spermatids (STD; 710 cells), Sertoli cells (SC; 9,488 cells), myoid cells (MYD; 3,531 cells), and Leydig cells (LY; 245 cells). There was an increase in the percentage of SPGC in Stra8-deficient testes at both P12 (28% vs. 14% in wild type) and P21 (17% vs. 10% in wild type) (Figures S2B and S2C). At P21, although spermatocytes were abundant and spermatids formed in wild-type testes, there were only a few spermatocytes (0.2% in Stra8-deficient testes vs. 73% in wild-type) in Stra8-deficient testes, with spermatids being completely absent (Figures S2A and S2D). Signature genes were enriched for distinct biological function and metabolic pathway gene ontology (GO) terms in different cell types (Figures S2E and S2F).
Loss of Stra8 results in up- and downregulated gene expression
Stra8 is expressed in germ cells during the narrow window of meiotic initiation. To identify changes associated with the loss of Stra8-induced gene expression, we selected germ cell transcriptomes (Ddx4+; 5,124 cells) from wild-type and Stra8-deficient mice for further analysis. Pooled germ cell transcriptomes from wild-type and Stra8-deficient cells were used to define nine clusters (Figure 1A). Based on known marker genes, these nine clusters were annotated as male germ cells at consecutive differentiation stages from SSCs (196 cells) to spermatid 1 (Std1; 218 cells) and spermatid 2 (Std2; 270 cells) through differentiating spermatogonia (Diff; 745 cells), preleptotene spermatocyte (PreL; 1,055 cells), leptotene spermatocyte (L; 622 cells), zygotene spermatocyte (Zygo; 107 cells), pachytene spermatocyte (Pachy; 606 cells), and diplotene spermatocyte (Diplo; 1,305 cells) (Figure 1B).
Figure 1. scRNA-seq of wild-type and Stra8-deficient germ cells at P12 and P21.
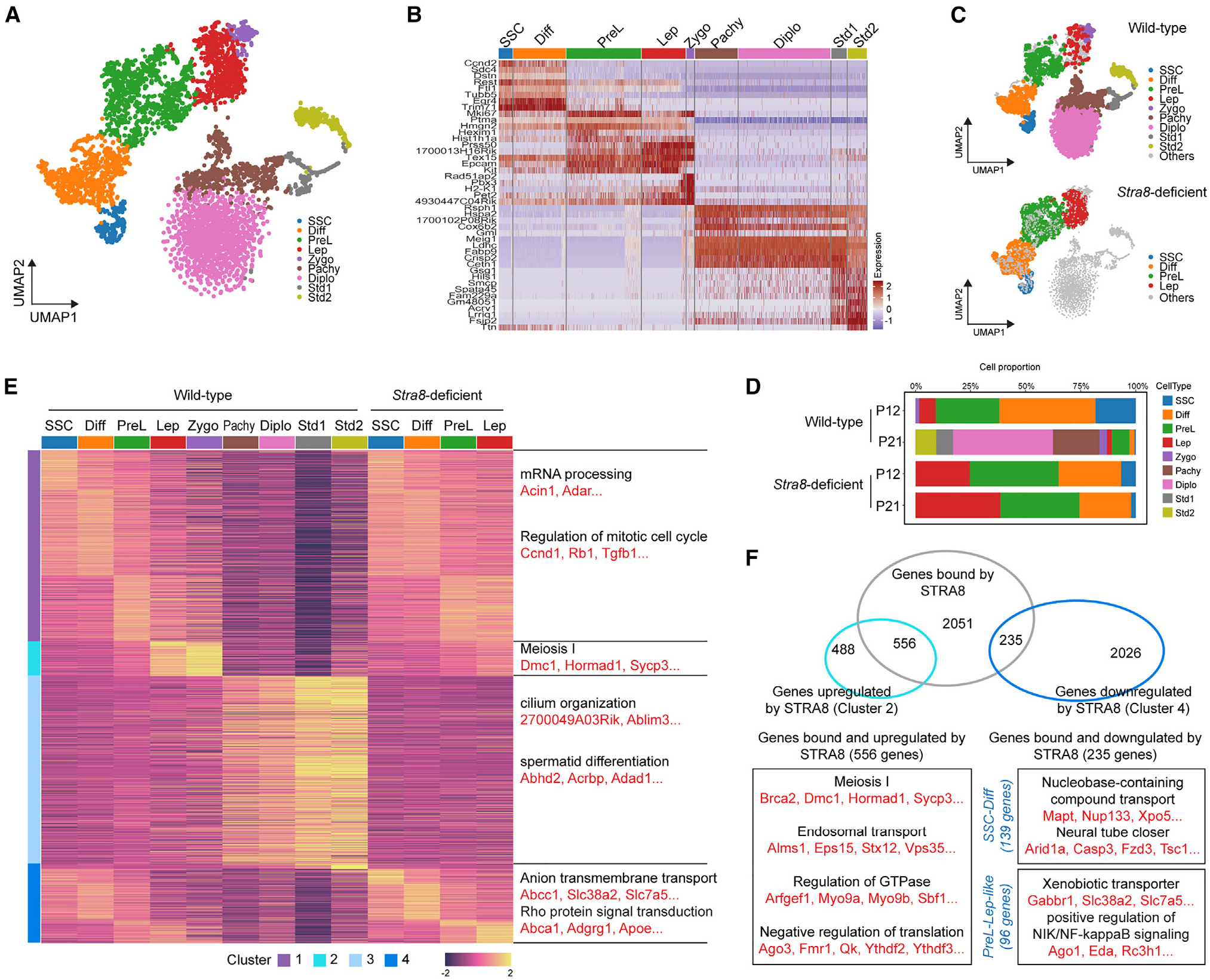
(A) Uniform manifold approximation and projection (UMAP) visualization of integrated projection from wild-type and Stra8-deficient germ cells at P12 and P21.
(B) Heatmap showing the expression of marker genes in each cluster.
(C) UMAP visualization of split projection by wild-type and Stra8-deficient germ cells.
(D) Bar plot showing the proportion of the different cell clusters at different time points.
(E) Clusters of genes across different cell types in wild-type and Stra8-deficient germ cells. Genes are grouped by activity cluster. Shown on the right are representative enrichments of biological processes.
(F) Venn diagrams with genes bound by Stra8 and representative genes that are upregulated and downregulated in wild-type and Stra8-deficient germ cells at the indicated stages.
See also Figures S1 and S2.
Consistent with their meiotic arrest phenotype, the transcriptomes of Stra8-deficient germ cells were arrested at the leptotene stage, the first stage of meiosis prophase (Figures 1C and 1D). Clustering analysis of differentially expressed genes showed four major clusters (Figure 1E). Genes in cluster 1 were expressed during early germ cells in wild-type germ cells from SSCs to PreL as well as in germ cells from Stra8-deficient germ cells at all stages. These genes are enriched for GO terms such as mRNA processing (e.g., Acin1, Adar) and the regulation of mitotic cell cycle (e.g., Ccnd1, Rb1). Genes in cluster 2 were only upregulated from wild-type leptotene to zygotene spermatocytes. As expected, they were enriched for GO terms related to meiosis, such as meiosis I (e.g., Dmc1, Hormad1, Sycp3). Of note, these meiosis-related genes were not upregulated in the leptotene spermatocytes of Stra8-deficient cells, which is consistent with their failed meiotic initiation and suggests that Stra8 is required for their activation. Thus, we consider that the “leptotene spermatocytes” in Stra8-deficient cells were not in meiosis but were “leptotene-like” cells. Genes in cluster 3 were only present in late meiotic (pachytene and diplotene) and postmeiotic (spermatid) wild-type cells. They were enriched in GO terms related to spermiogenesis, such as cilium organization (e.g., 2700049A03Rik, Ablim3) and spermatid differentiation (e.g., Abhd2, Acrbp, Adad1). Cluster 4 contained genes that were more highly expressed in the absence of Stra8, and they were enriched for GO terms related to nutrient transporters, such as anion transmembrane transporters (Abcc1, Slc38a2, Slc7a5). Together, these data suggest that Stra8 is required for both the upregulation (cluster 2) and downregulation (cluster 4) of gene expression during meiotic initiation.
A previous chromatin immunoprecipitation sequencing (ChIP-seq) study showed that Stra8 binds to the regulatory sequences of 2,841 genes.11 We found that, among the 1,044 genes whose upregulated expression during meiotic initiation requires Stra8 (cluster 2), Stra8 binds to 556 genes (53%) (Figure 1F). These genes are enriched for GO terms such as meiosis 1 (e.g., Brca2, Dmc1, Hormad1), endosomal transporter (e.g., Alms1, Eps15, Stx12), and regulation of guanosine triphosphatase (e.g., Arfgef1, Myo9a, Myo9b). Interestingly, among the 2,261 genes whose downregulated expression required Stra8 (cluster 4), Stra8 bound to 235 genes (10%) (Figure 1F). Since Stra8 is robustly expressed during meiotic initiation in preleptotene spermatocytes, we further narrowed down the list to 96 genes, whose aberrant upregulation occurs in preleptotene to leptotene-like spermatocytes in Stra8-deficient cells, suggesting that these genes that are repressed by Stra8. These genes are enriched in GO terms related to nutrient transporters such as “xenobiotic transporter” (e.g., Gabbr1, Slc38a2, Slc7a5) and “positive regulation of NIK/NF-kB signaling” (e.g., Ago1, Eda, Rc3h1). Interestingly, in SSCs and differentiating spermatogonia, we found that the loss of Stra8 also results in the upregulation of 139 Stra8-bound genes, whose GO terms were enriched for “nucleobase-containing compound transport” (e.g., Mapt1, Nup133, Xpo15) and “neural tube closer” (e.g., Arid1a, Casp3, Fzd3). As previously reported,16,17 a possible explanation for this observation is that Stra8 expression begins in differentiating spermatogonia. Thus, in addition to meiotic initiation, Stra8 has a regulatory role in spermatogonial differentiation.18 These data together suggest that Stra8 has a role as a transcriptional repressor during meiotic initiation.
Dynamic regulation of nutrient transporter gene expression by Stra8
There have been few descriptions of nutrient transporter gene expression in germ cells during spermatogenesis. To characterize this, we assembled a gene-expression map for most nutrient transporters19 by using our scRNA-seq dataset (Figure 2A). We found that mitotic germ cells (SSCs, differentiating spermatogonia, and preleptotene spermatocytes), meiotic germ cells (leptotene, zygotene, pachytene, and diplotene spermatocytes), and postmeiotic germ cells (spermatid 1 and spermatid 2) display distinctive patterns of nutrient transporter gene expression. In mitotic male germ cells, the nutrient transporter genes for lipids (e.g., Sar1b, Acat1), amino acids (e.g., Slc38a1, Slc3a2), inorganic solutes (e.g., Slc9a3r1), organic solutes (e.g., Slc5a3, Slc44a1), sugars (e.g., Slc2a1), metal ions (e.g., Slc39a7), and nucleotides (e.g., Slc25a36) were highly expressed, while the expression of nutrient transporter genes for bile salt, vitamins, and water was low. In meiotic germ cells, there appears to be global downregulation of nutrient transporter gene expression, except for that of the lipid transporter gene Abcq2, the vitamin transporter gene Rbp4, the inorganic solute transporter gene Slc4a7, the metal ion transporter genes Slc31a1 and Slc30a4, and the nucleotide transporter genes Slc35b2 and Slc35b3. Postmeiotic germ cells highly express the genes for nutrient transporters of water (Aqp7, Aqp11), amino acids (Slc25a39), organic solutes (Slco3a1), sugars (Slc2a5), metal ions (Slc39a8, Slc39a3), and nucleotides (Slc25a33).
Figure 2. Nutrient transporter gene expression in wild-type and Stra8-deficient germ cells.
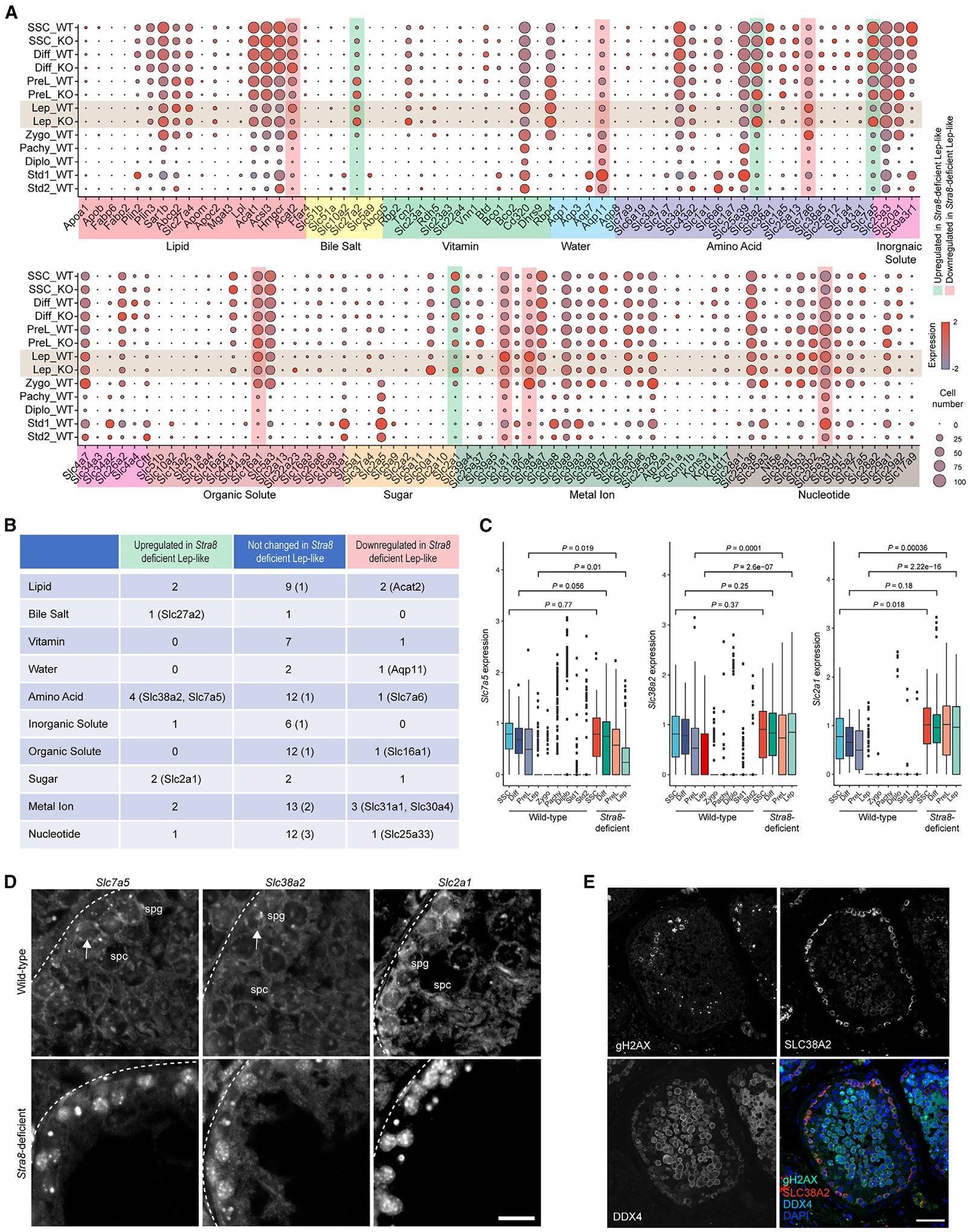
(A) Expression patterns of specific genes involved in nutrient absorption and transport in each cell type in wild-type and Stra8-deficient germ cells. Each dot represents a gene, of which the color saturation indicates the average gene-expression level and the size indicates the percentage of cells expressing the gene.
(B) Table summarizing nutrient transporter genes that are upregulated and downregulated genes in wild-type and Stra8-deficient germ cells at the leptotene stage. Genes bound by Stra8 are indicated in parentheses.
(C) Box plot showing the expression levels of Slc7a5, Slc38a2, and Slc2a1 at different stages of spermatogenesis in wild-type and Stra8-deficient germ cells (median ± SE). Statistical analyses between wild-type and Stra8-deficient samples in each cell type were performed using Student’s t test, with p values shown.
(D) ISH of Slc7a5, Slc38a2, and Slc2a1 mRNA in wild-type and Stra8-deficient testes. Scale bar, 20 μm.
(E) Staining of human testicular cross-sections using SLC38A2, γH2AX, VASA, and DAPI. VASA indicates germ cells. γH2AX indicates meiotic germ cells. Scale bar, 50 μm.
See also Figures S3 and S4.
Interestingly we found that at the leptotene stage, loss of Stra8 results in the upregulation of 13 nutrient transporter genes, among which Stra8 binds to four genes. These genes encode transporters for bile acid (Slc27a2), amino acid (Slc38a2, Slc7a5), and glucose (Slc2a1), suggesting that their expression is downregulated by Stra8 during meiotic initiation (Figures 2A and 2B). In addition, it appears that there is an increased number of cells in Stra8-deficient testes that express these nutrient transporters (Figure 2A). Slc27a2 is also known as fatty acid transport protein 2 (FATP2).20 Slc38a2, an amino acid transporter also known as Snat2, is a Na+-coupled net importer of neutral amino acids, particularly glutamine.21 Slc7a5, also known as Lat1, is a glutamine antiporter that imports large neutral amino acids, e.g., leucine, by exchanging with intracellular glutamine.22 Slc2a1, also known as Glut1, is a primary transporter for cellular glucose uptake.23 Meanwhile, loss of Stra8 results in the downregulation of ten nutrient transporter genes, suggesting that their expression is upregulated by Stra8 during meiotic initiation. Additionally, there were fewer cells in Stra8-deficient testes that express these nutrient transporters (Figure 2A). Among these genes, Stra8, together with Meiosin, an important cofactor for Stra8,12 binds to six genes. These genes encode transporters for water (Aqp11), amino acids (Slc7a6), organic solutes (Slc16a1), metal ions (Slc31a1, Slc30a4), and nucleotides (Slc25a33) (Figures 2A, 2B, and S3A). The only exception was the lipid transporter, Acat2, for which ChIP-seq results indicated only Stra8, but not Meiosin, binding (Figure S3A). These data suggest that the expression of these genes is upregulated by Stra8 during meiotic initiation.
Stra8 regulation of nutrient transporter expression
Nitrogen starvation drives meiotic fate decisions in yeast cells.4 Interestingly, glutamine is a major donor of nitrogen in mammalian metabolism.24 We show that the expression of the glutamine transporter genes Slc7a5 and Slc38a2, and the glucose transporter gene Slc2a1, is progressively downregulated during meiotic initiation (Figure 2C). Similar results were obtained when we investigated the bulk RNA-seq dataset25 and the slide-seq dataset26 reported by other groups (Figures S3B and S3C). In Stra8-deficient germ cells, the expression of Slc7a5, Slc38a2, and Slc2a1 was aberrantly sustained until the leptotene-like stage (Figure 2C). To confirm these results, we conducted in situ hybridization by RNAScope, which showed that, while Slc7a5, Slc38a2, and Slc2a1 were expressed in early germ cells near the basement membrane in wild-type testes, their expression was robustly upregulated in Stra8-deficient testes (Figure 2D). Consistently, our data show that in human testicular sections, SLC38A2 expression is detected only in mitotic spermatogonia and not in meiotic spermatocytes (Figure 2E), indicating that a rapid downregulation of SLC38A2 expression is associated with the onset of meiosis during human spermatogenesis.
To test whether Stra8 directly represses Slc7a5, Slc38a2, and Slc2a1 expression, we transiently transfected a full-length human STRA8-expressing plasmid into F9 embryonic carcinoma (EC) cells. Ectopic Stra8 expression inhibited Slc7a5, Slc38a2, and Slc2a1 expression at both the mRNA and protein levels (Figures 3A–3C). Similar results were obtained when we used F9 cells stably transfected with Stra8 (Figure S4A).
Figure 3. Regulation of nutrient transporter gene expression by Stra8.
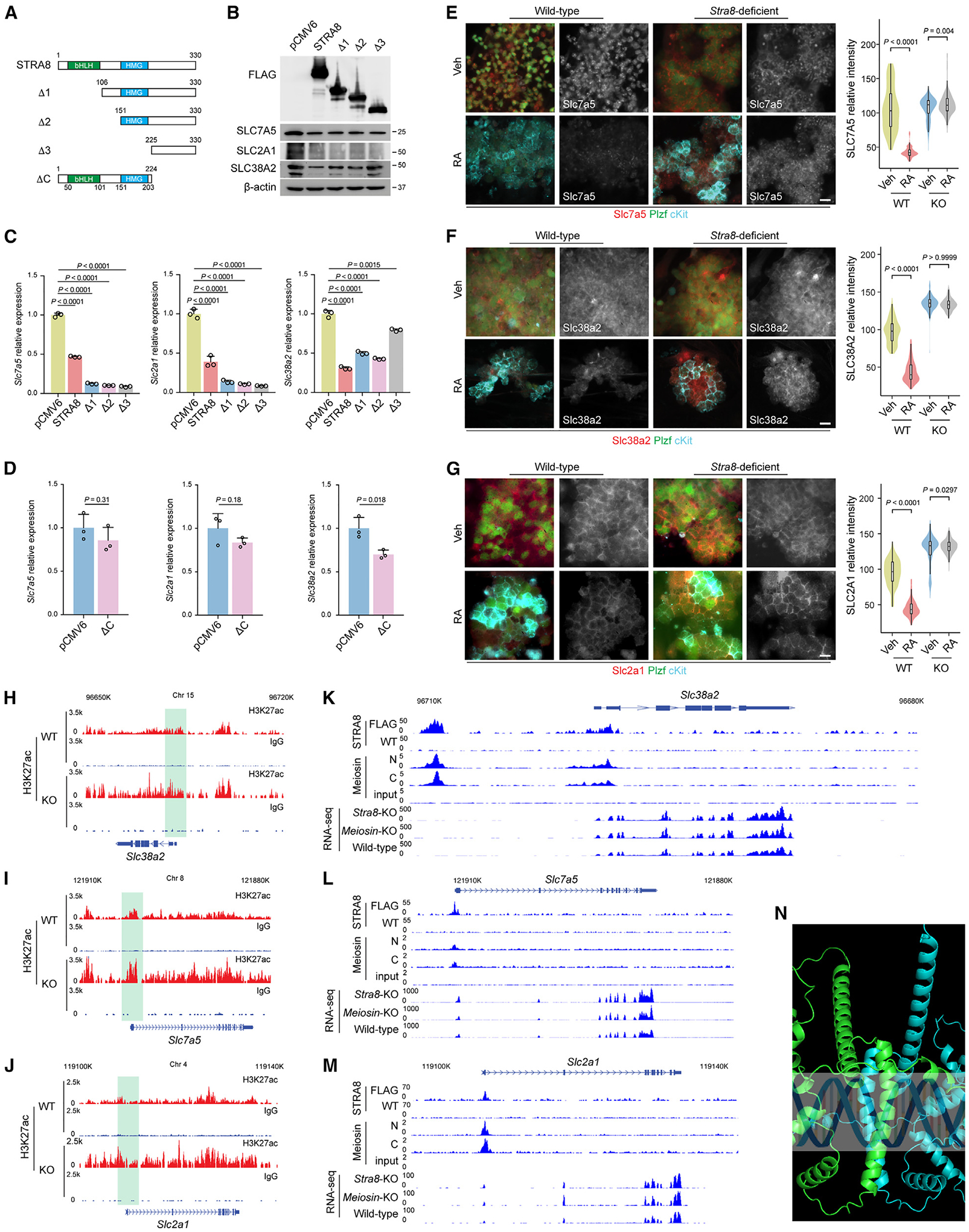
(A) Schematic representation of full-length Stra8 and its deletion constructs.
(B) The levels of Stra8 and its deletion constructs were detected by immunoblotting with an antibody against FLAG. Immunoblotting with antibodies against transporter proteins are shown.
(C and D) qPCR analysis of Slc7a5, Slc38a2, and Slc2a1 expression following transient transfection of STRA8 and its N terminus deletion mutants (C) and C terminus deletion mutant (D) into F9 cells for 48 h. Data represent mean ± SE. n = 3 independent transfections. Statistical analyses between full-length STRA8 and its N terminus deletion mutants (C) or its C terminus deletion mutant (D) were performed using Student’s t test, with p values shown.
(E–G) Representative immunofluorescence images of nutrient transporter protein Slc7a5 (E), Slc38a2 (F), and Slc2a1 (G) expression are shown in cultured wild-type (WT) and Stra8-deficient (KO) spermatogonial stem cells (SSCs) with or without RA treatment (100 nM) for 24 h. Scale bars, 10 μm. Violin plots of quantification results are shown on the right. p values were calculated by one-way ANOVA with Bonferroni’s multiple comparison test.
(H–J) Genome tracks of the Slc38a2 (H), Slc7a5 (I), and Slc2a1 (J) genes for CUT&RUN of H3K27ac using wild-type and Stra8-deficient testes at P12. In CUT&RUN of H3K27ac, samples incubated with preimmune immunoglobulin G were used as the negative controls.
(K–M) Genome tracks of the Slc38a2 (K), Slc7a5 (L), and Slc2a1 (M) genes for Stra8 ChIP-seq, Meiosin ChIP-seq, and RNA-seq with indicated genotypes. In Stra8-ChIP-seq, FLAG antibody was used to pull down Stra8 in FLAG-tagged Stra8 knockin mouse. Wild-type cells, in which Stra8 is not tagged, were used as the negative control.
(N) AlphaFold2 to predict the structure of the human STRA8 and MEIOSIN complex.
See also Figures S3–S5.
Human STRA8 contains a basic helix-loop-helix (bHLH) domain and a high-mobility group (HMG) domain11 (Figure 3A). A glutamic-acid-rich domain is present in mouse Stra8 but absent in human STRA8. To determine the domain crucial for Stra8 repression of Slc7a5, Slc38a2, and Slc2a1 expression, we first generated serial N terminus deletion mutants of Stra8 (Figure 3A). For Slc7a5 and Slc2a1, we found that the C terminus of Stra8 is essential for its inhibitory effect on Slc7a5 and Slc2a1, suggesting that neither the bHLH nor the HMG domain is required for this process. Moreover, deletion of the Stra8 N terminus (amino acids 1–105) that contains the bHLH domain potentiated Stra8 repression of these genes (Figures 3A–3C). For Slc38a2, however, deletion of both amino acids 1–105, which contains the bHLH domain, and amino acids 151–225, which contains the HMG domain, results in an elevated Slc38a2 expression, suggesting that both bHLH and HMG domains are important for Stra8 repression of Slc38a2 expression (Figures 3A–3C). We further generated a C terminus deletion mutant of Stra8 (ΔC) that contains the bHLH and HMG domains; this mutant failed to repress Slc7a5 and Slc2a1 but exhibited moderate but significant repression of Slc38a2 expression (Figure 3D).
Stra8 is induced by RA.27,28 We found that RA treatment (100 nM) for 24 h in cultured SSCs was sufficient to downregulate Slc7a5, Slc38a2, and Slc2a1 expression (Figure S4B).15 Consistently, our analysis of a dataset of newborn mice receiving RA treatment in vivo showed a similar effect29 (Figure S4C). To test this at the protein level, we showed that RA treatment induced upregulation of Stra8 expression (Figure S4D) and downregulation of Slc7a5 (43 ± 8 vs. 105 ± 32), Slc38a2 (44 ± 14 vs. 98 ± 16), and Slc2a1 (44 ± 12 vs. 94 ± 19) expression (Figures 3E–3G). These effects require Stra8, in that Stra8-deficient SSC cultures that received parallel RA treatment did not exhibit downregulation of the expression of these nutrient transporters, Slc7a5 (114 ± 19 vs. 107 ± 16), Slc38a2 (134 ± 8 vs. 135 ± 10), and Slc2a1 (131 ± 10 vs. 126 ± 21) (Figures 3E–3G).
By investigating a ChIP-seq dataset for RA and RA receptor (RAR) treatment in germ cells deposited in the Gene Expression Omnibus (GEO: GSE116798), we found that RAR does not bind to the regulatory sequences of Slc7a5, Slc38a2, or Slc2a1 (Figure S4E). Moreover, in these cells we examined two active histone marks, H3K27ac and H3K4me3.30,31 Our data show that, while RA reduces the H3K27ac of the nutrient transporter genes that are downregulated by Stra8 (Figure S4F), RA induces the H3K27ac of the nutrient transporter genes that are upregulated by Stra8 (Figure S4G); this result suggests that H3K27ac is sensitive to RA-Stra8 signaling and its regulation of target gene expression. However, for H3K4me3 our data show that, irrespective of the nutrient transporter genes that are upregulated or downregulated by Stra8, RA treatment decreased H3K4me3, suggesting that H3K4me3 is specific to Stra8 (Figure S3). Thus, to determine whether Stra8 is responsible for these epigenetic changes, we further conducted cleavage under targets and release using nuclease (CUT&RUN) assays for H3K27ac employing juvenile testicular cells from wild-type and Stra8-deficient mice, followed by high-throughput genomic sequencing. Our results show that H3K27ac is upregulated in Stra8-deficient mice, indicating that Stra8 is required for the removal of H3K27ac marks in the Slc7a5, Slc38a2, and Slc2a1 genes, which are downregulated by Stra8 (Figures 3H–3J). Reciprocally, our data show that the H3K27ac of Acat2, Slc31a1, and Slc30a4 was downregulated in Stra8-deficient cells, suggesting that Stra8 upregulates their expression through H3K27ac (Figure S5A).
A previous study identified Meiosin as an important partner for Stra8.12 Our RT-PCR results detected low levels of Meiosin in cultured SSCs and F9 EC cells, which were upregulated by RA treatment (Figure S5B). We checked the ChIP-seq results for Meiosin.12 Our results show that Meiosin binds to Slc7a5, Slc38a2, and Slc2a1 where Stra8 binds (Figures 3K–3M). For Slc2a1 and Slc7a5, the Stra8-Meiosin complex binds at the transcription start site (TSS) (Figures 3L and 3M). Interestingly, for Slc38a2, the Stra8-Meiosin complex binds not only to the TSS but also to a site that is approximately 10 kbp upstream of the TSS, suggesting that Slc38a2 is subject to additional transcriptional regulation (Figure 3K). This is consistent with our observation that, while Stra8 C terminus (amino acids 225–330) is required for downregulation of Slc7a5 and Slc2a1 expression, Stra8 amino acids 151–224 further inhibit Slc38a2 expression (Figure 3A). To examine the regulation of Slc38a2, Slc7a5, and Slc2a1 expression by Meiosin, we analyzed the published RNA-seq dataset,12 which showed that the loss of Meiosin results in a moderate upregulation of Slc38a2, Slc7a5, and Slc2a1 expression that is less profound than that following loss of Stra8 (Figures 3K–3M). These data suggest that downregulated expression of Slc38a2, Slc7a5, and Slc2a1 during meiotic initiation is highly dependent on Stra8.
We also used AlphaFold2 to predict the structure of the STRA8 and MEIOSIN complex. Residues 50–101 from STRA8 and residues 62–116 from MEIOSIN are predicted to form an HLH-domain shape consisting of a helix-loop-helix, which folds into two heterodimers that are oriented in a head-to-tail configuration resulting in an antiparallel four-helix bundle (Figure 3N).
RA inhibits glutamine and glucose uptake via Stra8
We next evaluated whether Stra8 regulates nutrient uptake in germ cells. RA treatment in wild-type SSCs resulted in a reduction of glucose (40 ± 7 vs. 81 ± 29) and glutamine (70 ± 31 vs. 144 ± 61) uptake; however, these effects for glucose (105 ± 40 vs. 90 ± 20) and glutamine (135 ± 21 vs. 135 ± 38) were not observed in Stra8-deficient SSCs (Figures 4A and 4B), suggesting that Stra8 is required for RA-mediated inhibition of glutamine and glucose uptake. A similar effect in glucose uptake activity was also observed from freshly isolated undifferentiating (c-Kit-negative integrin-α6-high) and differentiating (c-Kit-negative integrin-α6-low) male wild-type and Stra8-deficient germ cells, in that only wild-type, but not Stra8-deficent, germ cells exhibited a profound decrease in glucose uptake upon their meiotic differentiation (Figure S5C). There was also a slight reduction in glucose uptake in Stra8-deficent c-Kit positive cells, suggesting that, in addition to Stra8, other factors could inhibit glucose uptake during germ cell differentiation. Reciprocally, our data show that ectopic Stra8 expression is sufficient to limit glutamine and glucose uptake (Figures 4C and 4E). Consistently, Mito Fuel Flex analyses showed that ectopic Stra8 expression significantly compromised the flexibility and capacity of cells to utilize glucose and glutamine, but not fatty acids, for fuel utilization (Figures 4F–4H).
Figure 4. Stra8 regulates glutamine and glucose uptake.
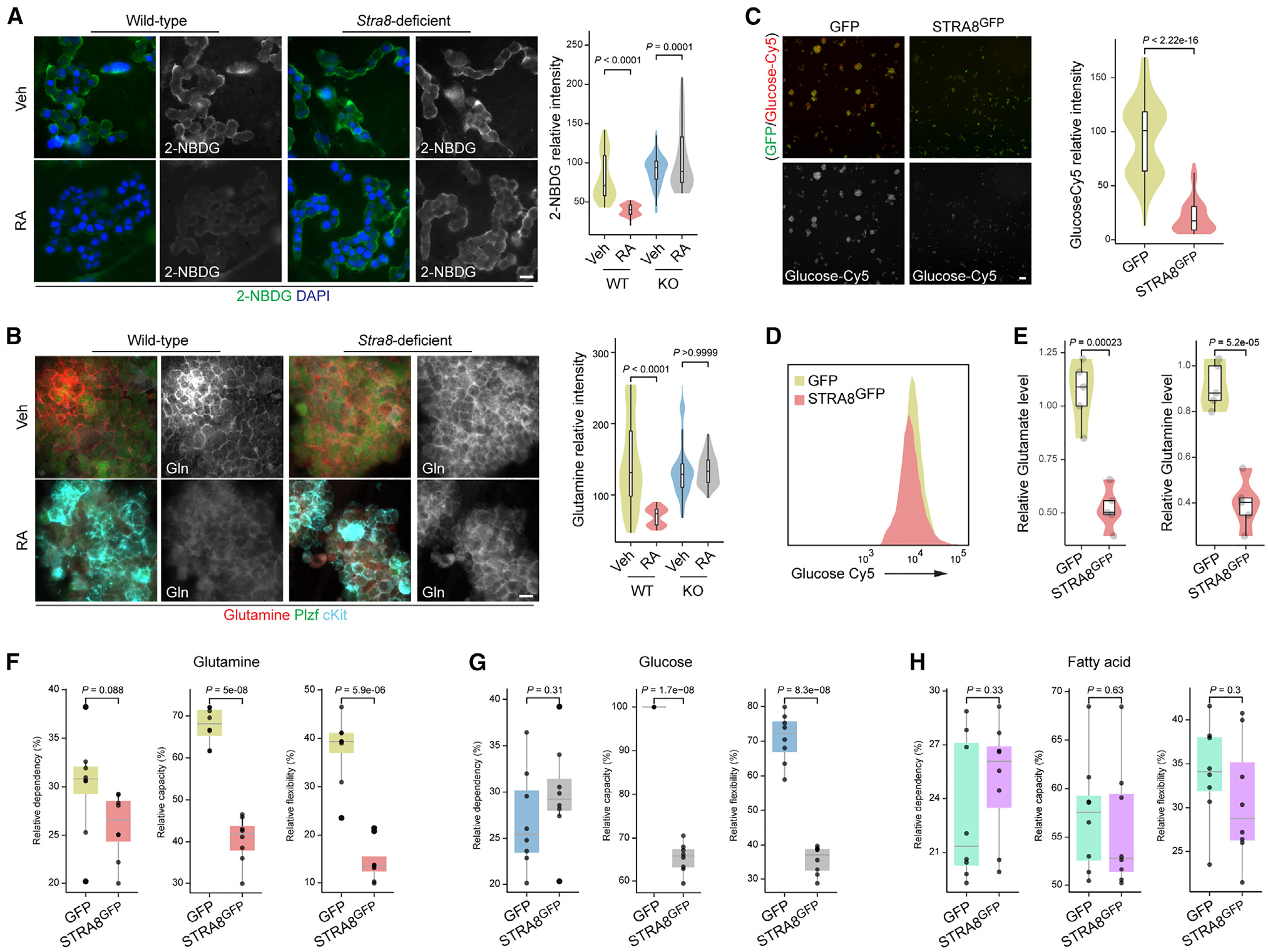
(A) The fluorescence of 2-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-2-deoxyglucose (2-NBDG) was measured, with representative statistical plot profiles of cultured wild-type (WT) and Stra8-deficient SSCs with or without RA treatment (100 nM) for 24 h. Representative images from three independent experiments. Scale bar, 10 μm. Violin plots of quantification results are shown on the right. p values were calculated by one-way ANOVA with Bonferroni’s multiple comparisons test.
(B) Representative immunofluorescence of glutamine staining detected, with representative statistical plot profiles of cultured wild-type and Stra8-deficient SSCs with or without RA treatment (100 nM in DMSO) for 24 h. Representative images from three independent experiments. Gln, glutamine. Scale bar, 10 μm. Violin plots of quantification results are shown on the right. p values were calculated by one-way ANOVA with Bonferroni’s multiple comparison test.
(C) Fluorescence of glucose-Cy5 staining detected and representative violin plot profiles of F9 cells transfected with GFP (control) and Stra8GFP expression plasmids. Representative images from three independent experiments. Scale bar, 10 μm. Violin plots of quantification results are shown on the right. p value was calculated by Student’s t test.
(D) FACS plot for detection of glucose-Cy5 and representative plot profiles of F9 cells transfected with expression plasmids for GFP (control) and Stra8GFP.
(E) Glutamate and glutamine levels were measured in cultured F9 cells transfected with GFP (control) and Stra8GFP expression plasmids. Violin plots of quantification results are shown on the right. p values were calculated by Student’s t test.
(F–H) Box plots of Mito Fuel Flex test profile for substrate fuel dependency, and the three major fuel pathways of glutamine (F), glucose (G), and fatty acid (H). p values were calculated by Student’s t test.
See also Figure S5.
Slc38a2 knockdown upregulates meiosis-related gene expression
Our data suggest that the downregulation of nutrient transporter genes is associated with meiotic gene activation and initiation in mammalian germ cells. Previously, we compiled a list of 165 meiosis-related genes whose expression is sensitive to nutrient restriction signals and RA.15 We tested their correlation with these nutrient transporter genes in the Genotype-Tissue Expression (GTEx) database.32 Our data showed that Slc7a5 and Slc2a1 had moderate negative correlations with 7- and 11-meiosis-related genes, respectively (Pearson correlation between −0.6 and −0.3) (Figures 5A and 5B). Intriguingly, Slc38a2, which mediates neutral amino acid uptake (primarily glutamine), exhibited strong negative correlations with 26 genes (Pearson correlation less than −0.6) and moderate and negative correlations with 68 genes (Pearson correlation between −0.6 and −0.3) (Figures 5A and 5C). These genes included many genes essential for meiosis, e.g., Psmc3ip (encoding Hop2), Sycp3, Sycp1, and Hormad1. Of note, all genes that exhibit moderate negative correlations with Slc2a1 were negatively correlated with Slc38a2, suggesting potential pathway overlap.
Figure 5. Nutrient transporters regulate meiotic gene expression.
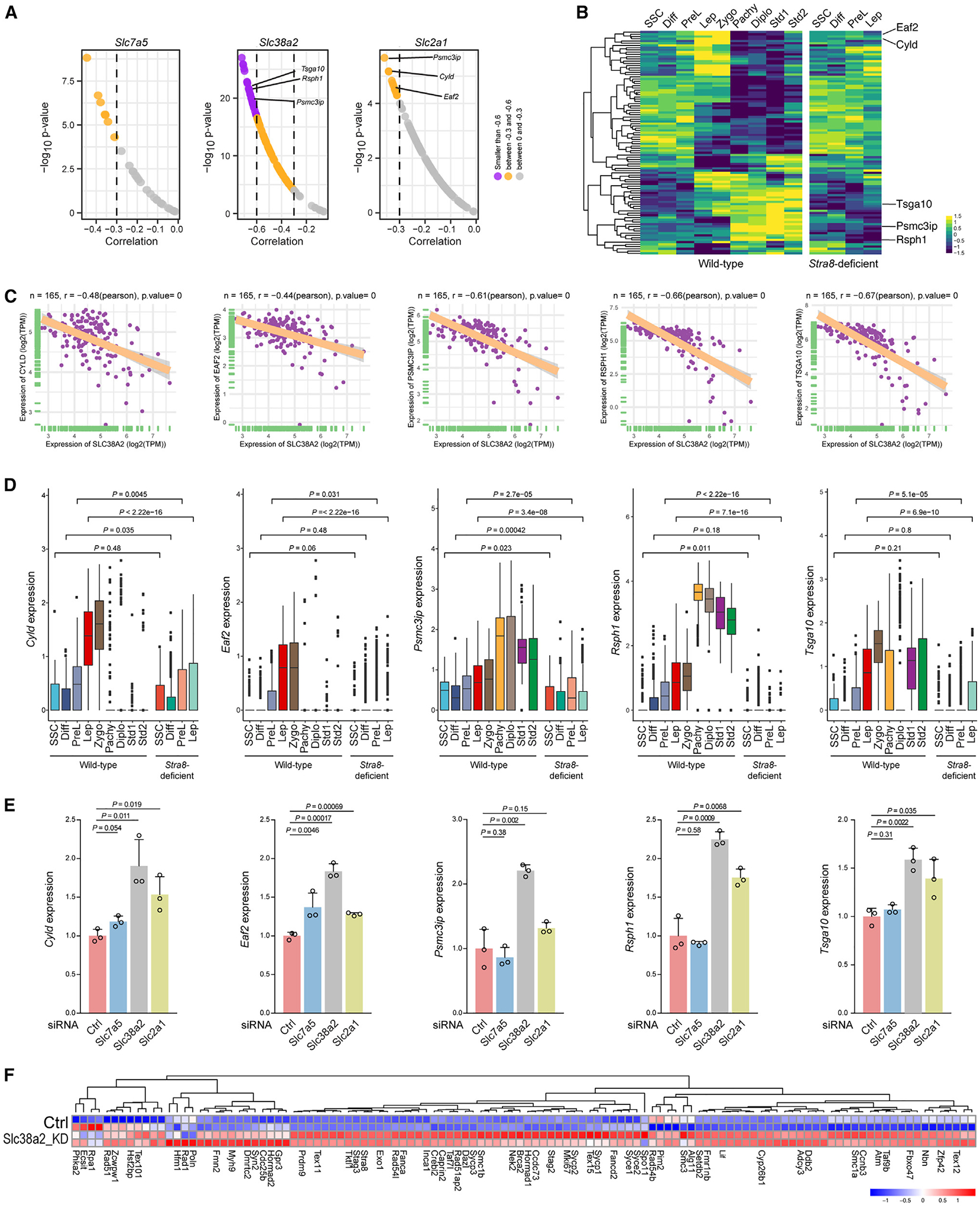
(A) Top correlation of representative transporter genes (Slc7a5, Slc38a2, and Slc2a1) with selected meiotic genes in 165 testis samples in the GTEx datasets.
(B) Heatmap of the expression of meiotic genes that show strong or moderate correlation with nutrient transporter genes from the GTEx datasets in wild-type and Stra8-deficient germ cells. The top five meiotic genes that are negatively correlated with Slc38a2 and Slc2a1 and are decreased in Stra8-deficient germ cells are shown on the right.
(C) GTEx datasets showing the correlation of Slc38a2 with selected meiotic genes in 165 testis samples. The linear regression curve is demonstrated. The 95% confidence interval (shaded area) is shown in each panel. The range of confidence intervals is provided in the source data. Two-tailed t-statistic p value and coefficient (R) of Pearson’s correlation are shown at the top.
(D) Box plots showing the expression level of representative meiotic genes in each cluster from wild-type and Stra8-deficent germ cells. Statistical analyses between wild-type and Stra8-deficient samples in each cell type were performed using Student’s t test, with p values shown.
(E) qPCR showing the expression level of representative meiosis genes after Slc7a5, Slc38a2, and Slc2a1 knockdown in mouse SSC cultures. Data are median ± SE. Statistical analyses between samples with control siRNA and siRNA against Slc7a5, Slc38a2, and Slc2a1 were performed using Student’s t test, with p values shown.
(F) Heatmap of meiosis-related gene expression from control or Slc38a2 knockdown (Slc38a2 KD). Genes shown were selected from those that exhibited both a negative correlation with Slc38a2 in the GTEx dataset (A) and upregulation in Slc38a2 KD samples detected by RNA-seq.
See also Figure S6.
When examining the expression pattern of these genes, we found that many genes are upregulated upon entry into meiosis in wild-type germ cells but not in Stra8-deficient germ cells, which also exhibit aberrant upregulation of nutrient transporter gene expression (Figures 2 and 5). We selected Cyld, Eaf2, Psmc3ip (also known as Hop2), Rsph1, and Tsga10 as examples, because our data show that these genes are robustly activated during meiotic initiation in wild-type germ cells; however, the expression of these genes during meiotic initiation is significantly impaired in Stra8-deficient germ cells that fail to enter meiosis (Figures 5C and 5D). Among these genes the most characterized is Psmc3ip, which encodes Hop2. Psmc3ip was first identified in yeasts and was found to be essential for synaptonemal complex formation.33 Later, Hop2-knockout mice showed profound meiotic defects in synapsis and meiotic double-strand break repair.34 To directly test whether the downregulation of nutrient transporter genes is sufficient to induce meiotic gene activation, we transfected small interfering RNA (siRNA) against Slc7a5, Slc38a2, and Slc2a1 (Figures 5E and S6). Consistent with GTEx analysis (Figure 5A) we found that, while knockdown of Slc7a5 only induced Eaf2 expression, knockdown of Slc38a2 significantly induced the expression of all five of these meiotic genes, and knockdown of Slc2a1 significantly but less potently induced the expression of four genes, excluding Psmc3ip.
Because the GTEx datasets show that Slc38a2, but not Slc7a5 or Slc2a1, exhibits a negative correlation with most meiotic genes, we conducted RNA-seq for cultured SSCs with control siRNA and siRNA against Slc38a2 (Figure 5F). We showed that, among the 82 meiotic genes that exhibited moderate and strong negative correlations with Slc38a2, Slc38a2 knockdown is sufficient to induce the upregulation of 51 genes (62%). Thus, these data from multiple independent approaches, including the GTEx dataset, qRT-PCR, and RNA-seq, support the hypothesis that Slc38a2 plays a role in negatively regulating meiosis-related gene expression.
mTORC1 and PKA activities link Slc38a2 with meiosis-related gene expression
Mammalian target of rapamycin complex 1 (mTORC1) and protein kinase A (PKA) are major nutrient sensors that control meiotic fate decisions in yeasts.2,14 Nutrient transporter expression, e.g., Slc38a2, often activates mTORC1 activity. We first examined mTORC1 and PKA activities in Stra8-deficient testes, where the nutrient transporter genes Slc7a5, Slc38a2, and Slc2a1 are highly expressed (Figure 2). For mTORC1 activity, we used phosphorylated ribosomal protein S6 kinase (p-S6K; Thr398), phosphorylated ribosome protein S6 (p-rpS6; Ser235/236), phosphorylated eukaryotic translation initiation factor, and 4E-binding protein (p-4EBP1; Thr37/46). For PKA activity, we used phosphorylated cyclic AMP response element-binding protein (CREB) (p-CREB; Ser133). Our data show that mTORC1 and PKA activities are significantly upregulated in germ cells of Stra8-deficient testes (Figures 6A and 6B). To determine the nutrient transporter gene(s) that was responsible for the increased mTORC1 and PKA activities, we used siRNAs to knock down Slc7a5, Slc38a2, and Slc2a1 (Figure S6A). We found that knockdown of Slc38a2 and Slc2a1 resulted in the downregulation of mTORC1 activity, and knockdown of Slc7a5 with Slc38a2 reversed p-S6K/S6K expression (Figure 6C). Furthermore, knockdown of Slc38a2, but not Slc7a5 and Slc2a1, decreased PKA activity (Figures 6D and S6B).
Figure 6. Nutrient transporters regulate mTORC1 and PKA activities.
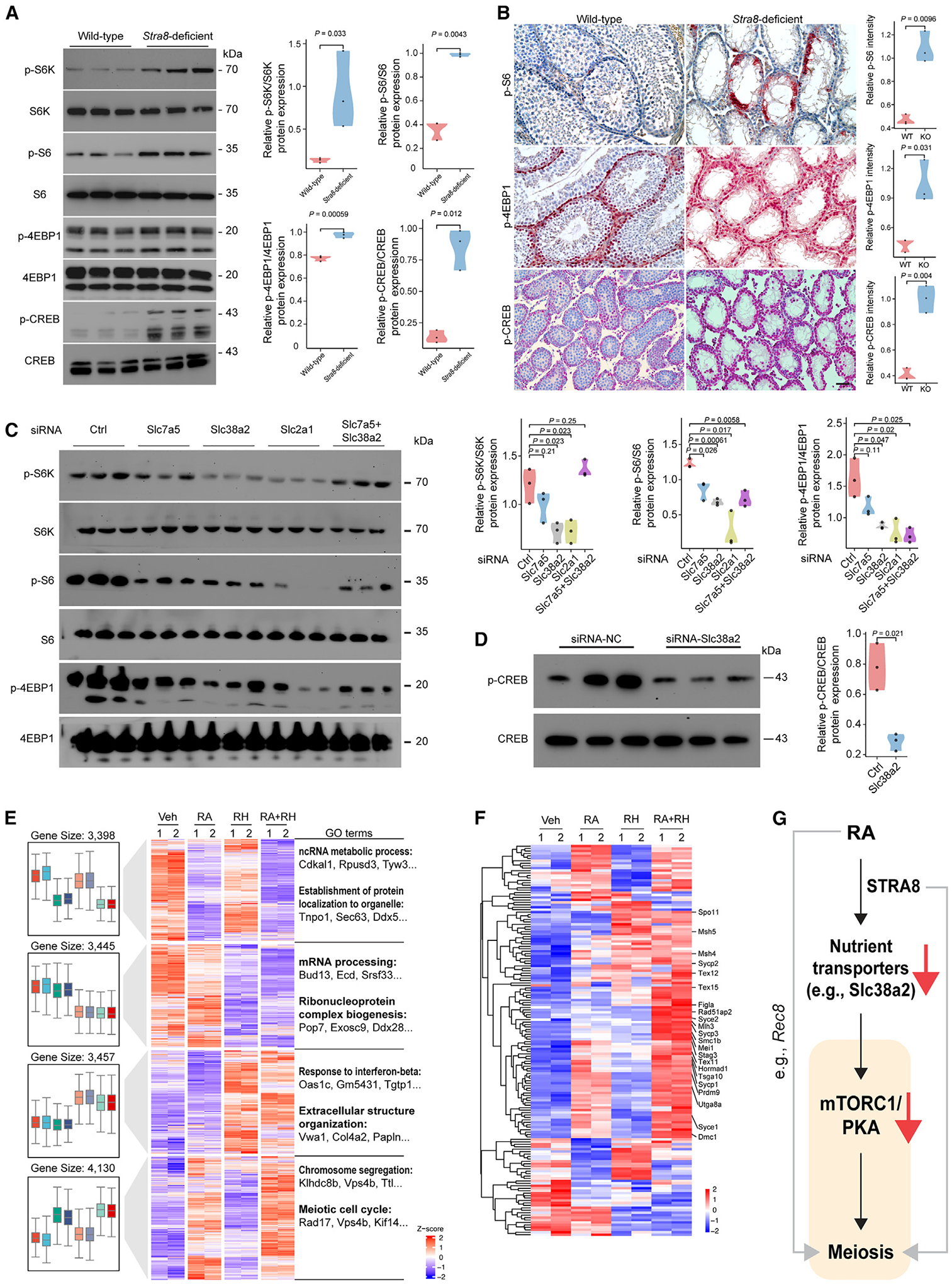
(A) Representative immunoblotting analysis using total testicular lysates from wild-type and Stra8-deficient mice at P21. Right: violin plots of indicated phosphorylated proteins normalized to total proteins (n = 3). p values were calculated by Student’s t test.
(B) Representative images of p-S6, p-4EBP1, and p-CREB immunohistochemistry staining. Scale bars, 100 μm. Right: violin plots showing staining intensity of indicated proteins (n = 3). p values were calculated by Student’s t test.
(C) Representative immunoblotting analysis using total lysates from Slc7a5, Slc38a2, Slc2a1, and Slc7a5+Slc38a2 knockdown cells. Right: violin plots of indicated phosphorylated proteins normalized to total proteins (n = 3). Statistical analyses between samples with control siRNA and siRNA against Slc7a5, Slc38a2, Slc2a1, and Slc7a5+Slc38a2 were performed using Student’s t test, with p values shown.
(D) Representative immunoblotting analysis using total lysates from Slc38a2 knockdown cells. Right: violin plots of indicated p-CREB normalized to total CREB (n = 3). p value was calculated by Student’s t test.
(E) Left: Unsupervised hierarchical clustering (UHC) and heatmap of gene expression in SSC cultures with indicated treatments for 2 days. Right: Top GO enrichments with representative genes in each cluster. Rapamycin was used at 100 nM in DMSO. H89 was used at 1 μM in DMSO.
(F) UHC and heatmap for the expression of 165 early meiosis-associated genes with the indicated treatment for 2 days.
(G) Schematic model that shows chordate morphogen RA inducing meiosis by silencing nutrient transporter expression, which in turn elicits a conserved nutrient restriction signal that involves TORC1 and PKA.
See also Figure S6.
To test whether Slc38a2 knockdown induces meiotic gene expression through mTORC1 and PKA inactivation, we inhibited mTORC1 using rapamycin and PKA using H89. We found that short-term treatment of rapamycin and H89 with RA was sufficient to induce the expression of key meiotic genes, including Dmc1, Hormad1, and Hop2 (Figures S6C and S6D). To assess the extent of gene-expression changes in an unbiased manner, we further conducted RNA-seq analysis. Our results show that treatment of rapamycin and H89 with RA induced the expression of a cluster of genes enriched for the GO functions “chromosome segregation” and “meiotic cell cycle” (Figure 6E). Neither treatment of rapamycin and H89 nor RA alone had this effect (Figure 6E). Further analysis showed that, among the 160 meiosis-related genes, treatment of rapamycin and H89 with RA was sufficient to induce 113 genes (71%) (Figure 6F). Together, these data suggest that downregulation of nutrient transporter genes (e.g., Slc38a2) activates meiotic gene expression in part through mTORC1 and PKA inactivation (Figure 6G).
DISCUSSION
Our recent study showed that nutrient restriction works with RA to induce meiotic inhibition in cultured male SSCs.15 Here our study revealed a dramatic downregulation of nutrient transporter genes during meiotic initiation, which is mediated by RA through Stra8. Our study suggests a novel, cell-autonomous mechanism for mammalian germ cells to generate a conserved signal (nutrient restriction) for meiotic fate determination in response to a chordate morphogen, RA. Thus, while yeast cells passively depend on environmental nutrient restriction to induce their meiosis, mammalian cells have devised a mechanism of self-starvation by withdrawing the expression of their nutrient transporters.
Our data revealed that a subset of the Stra8-deficient cell population should be classified as leptotene or, more precisely, leptotene-like, in terms of gene expression. In previous studies,9,10 the conclusion that Stra8-deficient germ cells are arrested at the preleptotene stage is because leptotene cells were not observed at morphological or cytological level. However, it should be noted that cells expressing Sycp3 are readily observed in Stra8-deficient testes, although Sycp3 is not properly loaded onto the chromosomes.12,35 Notably, the Stra8-deficient phenotype is not strain dependent, as we have observed the phenotype in both C57BL/6 inbred and C57BL/6XDBA2 F1 hybrid backgrounds.15 Together these data suggest that Stra8-deficient germ cells do not simply arrest at the preleptotene stage. Here our scRNA-seq analysis provides greater detail, as it allows us to analyze the Stra8-deficient phenotype at the transcriptomic level. Our data support the hypothesis that Stra8-deficient germ cells progress to a leptotene-like stage. Our results show that Stra8-deficient germ cells reach a stage that is generally comparable to that of wild-type leptotene cells, as evidenced by being in the same cluster on uniform manifold approximation and projection (UMAP) by unsupervised clustering. However, while wild-type cells in this cluster have started to express meiosis-related genes (e.g., Dmc1), Stra8-deficient cells do not. Thus, we consider that, while wild-type cells are “leptotene,” as leptotene is the first stage of meiosis prophase I, Stra8-deficient cells are “leptotene-like.” Thus, we annotated the data accordingly. It should be noted that this unsupervised clustering outcome—that is, wild-type and Stra8-deficient cells belong to the same cluster on UMAP while exhibiting differential meiosis-related gene expression—reflects the fact that the expression profile of meiosis-related genes represents a fraction of the overall transcriptome when cells are computed for clustering. Meanwhile, these data provide transcriptomic characterization of the Stra8-deficient phenotype at the single-cell resolution as follows: although the cells do not exhibit morphological or cytological features of leptotene, their transcriptomes have in part reached the leptotene stage.
To examine the cell-cycle difference between wild-type leptotene cells and Stra8-deficient leptotene-like cells, we used Ccnb1 and Top2a as representatives (Figure S6E). Ccnb1 is expressed in mitotic spermatogonia and deletion of Ccnb1 disrupts their proliferation and leads to male sterility.36 In contrast, during the cell cycle, Top2a is maximally expressed in the G2/M phase.37 During spermatogenesis, Top2a is highly expressed at the leptotene to pachytene stages.38 Consistent with these reports, our scRNA-seq data show that wild-type leptotene cells exhibit downregulated Ccnb1 but upregulated Top2a expression (Figure S6E). However, Stra8-deficient leptotene-like cells had sustained Ccnb1 expression, and Top2a expression was not activated (Figure S6E). These data suggest that Stra8-deficient leptotene-like cells have not properly transitioned from the mitotic to the meiotic cell cycle.
If RA induces downregulation of glutamine transporter expression to limit glutamine uptake, why is RA not sufficient to induce meiosis in complete SSC medium but needs to work in nutrient restriction medium?15 We hypothesize that the formulation of the nutrient-rich complete SSC medium is designed to promote rapid SSC growth in cultures but does not represent the condition where germ cells reside in vivo. For instance, while the plasma concentration of glutamine, as the most abundant amino acid, is 0.6–0.9 mM,39 the complete SSC medium contains 2 mM glutamine. Additionally, meiosis takes place in germ cells that reside behind the BTB, an ultrastructure known to limit nutrient flow,40,41 resulting in a lowered nutrient concentration. Thus, when SSCs are grown in the complete SSC medium, RA-induced downregulation of endogenous glutamine transporter expression may not be sufficient to elicit the nutrient restriction signal necessary for meiosis. In support of this hypothesis our data show that, compared to SSCs cultured in complete (nutrient-rich) SSC medium, SSCs cultured in the nutrient restriction medium exhibited a greater reduction in mTORC1 activity in response to RA treatment (Figure S6F).
Our results show that Stra8 downregulates Slc38a2 expression, which in turn inactivates mTORC1/PKA. To test whether mTORC1/PKA could affect Slc38a2 and Stra8 expression, our data showed that mTORC1/PKA inactivation modestly upregulated their expression (Figure S6G). Thus, these data confirm our proposed model in Figure 6G, which shows that downregulation of nutrient transporter expression is an upstream event leading to mTORC1/PKA inactivation and meiotic gene activation.
Intriguingly, germ cells of Stra8-deficient testes exhibited hyperactive mTORC1 activity (Figures 6A and 6B). While active mTORC1 usually switches off cellular autophagy, our past study shows that Stra8-deficient germ cells, on the contrary, exhibit robust autophagy activation.13 A possible explanation for this phenomenon could be that, in addition to elevated Slc38a2 expression, autophagy-generated nutrients through recycled cellular organelles and proteins also fuel mTORC1 activity.
Unlike in yeast cells,2 we found that mTORC1 and PKA inhibition was not sufficient to induce meiotic gene activation and that RA was required (Figure 6E). Thus, it is plausible that the regulation of meiotic fate decisions in mammalian germ cells is more complicated than that in yeasts. For instance, Stra8 has been shown to bind to the regulatory sequences of meiotic genes.11,12 In a separate study, we found that Stra8 is required to open the chromatin regions of meiotic genes using ATAC-seq (assay for transposase-accessible chromatin with sequencing) during meiotic initiation (unpublished observations). Thus, Stra8 could be a prime factor for meiotic genes that is necessary for their activation in response to the conserved nutrient restriction pathway controlled by mTORC1 and PKA.
Limitations of the study
Our current study is focused on meiosis-initiating germ cells. In SSCs and Diff spermatogonia, loss of Stra8 results in the upregulation of Stra8-bound genes, which are relevant to “nucleobase-containing compound transport” and “neural tube closer” (Figure 1). It has been reported that in mice lacking Stra8, undifferentiated spermatogonia accumulate in unusually high numbers after birth, whereas differentiating spermatogonia are depleted.18 Thus, while our current study is focused on meiotic initiation in PreL and L cells, the biological relevance of those downregulated genes in SSCs and Diff cells remains to be determined. Technically, F9 cells, which are EC cells, were used in some experiments (Figures 3C, 3D, 4C, and 4E). F9 cells are neither SSCs/spermatogonia nor spermatocytes. F9 cells were used in our studies for their robust Stra8 expression upon RA treatment as well as a tool cell to dissect the mechanism by which STRA8 inhibits nutrient transporter expression due to efficiency for transfection.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Additional information and requests for reagents and resources should be directed to and will be fulfilled by the lead contact, Ning Wang (nwang2@kumc.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
FastQ files of single-cell RNA-seq data have been deposited at the Gene Expression Omnibus (GEO) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PLZF | Santa Cruz | Cat# sc-28319; RRID:AB_2218941 |
| KIT | R&D Systems | Cat# AF1356; RRID:AB_354750 |
| SLC7A5 | Cell Signaling Technology | Cat# 5347S; RRID:AB_10695104 |
| SLC38A2 | Sigma Aldrich | Cat# HPA035180; RRID:AB_10669901 |
| SLC2A1 | Abcam | Cat# ab115730; RRID:AB_10903230 |
| gH2AX | Millpore | Cat# 05–636; RRID:AB_309864 |
| VASA | R&D Systems | Cat# AF2030; RRID:AB_2277369 |
| P70S6K | Cell Signaling Technology | Cat# 9234P; RRID:AB_2269803 |
| p-70S6K | Cell Signaling Technology | Cat# 9202S; RRID:AB_331676 |
| S6 | Cell Signaling Technology | Cat# 4858P; RRID:AB_916156 |
| p-S6 | Cell Signaling Technology | Cat# 2217S; RRID:AB_331355 |
| 4EBP1 | Cell Signaling Technology | Cat# 2855P; RRID:AB_560835 |
| p-4EBP1 | Cell Signaling Technology | Cat# 9644T; RRID:AB_2097841 |
| FLAG | Sigma Aldrich | Cat# F1804; RRID:AB_262044 |
| a6 integrin | BD Biosciences | Cat# 555736; RRID:AB_396079 |
| KIT | BD Biosciences | Cat# 553356; RRID:AB_398536 |
| donkey anti-mouse IgG Alexa Flour 488 | Life Technologies | Cat# A21202; RRID:AB_141607 |
| donkey anti-rabbit IgG Alexa Flour 568 | Life Technologies | Cat# A10042; RRID:AB_2534017 |
| donkey anti-goat IgG Alexa Flour 647 | Life Technologies | Cat# A21447; RRID:AB_141844 |
| Biological samples | ||
| Human testis testicular tissue from organ donors | Center for Organ Recovery and Education | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Superscript II Reverse Transcriptase | Thermo Fisher Scientific | Cat# 18080051 |
| NuPage 4 %–12 %Bis-Tris Protein Gel | Thermo Fisher Scientific | Cat# NP0322BOX |
| cOmplete, EDTA-Free | Roche | Cat# 4 693 132 |
| RIPA Buffer | Thermo | Cat# 89900 |
| all-transRA | Sigma-Aldrich | Cat# R2625 |
| 2-NBDG | Sigma-Aldrich | Cat# 72987 |
| Glucose-Cy5 | Sigma-Aldrich | Cat# SML3233 |
| collagenase IV | Thermo Fisher Scientific | Cat# 17104019 |
| TRIzol | Invitrogen | Cat# 15596018 |
| Power SYBR Green Master Mix | Applied Biosystems | Cat# A25742 |
| Lipofectamine 3000 | Invitrogen | Cat# L3000015 |
| N2 | R&D Systems | Cat# AR-009 |
| Pyruvic Acid | Sigma-Aldrich | Cat# P2256 |
| D-(+)-glucose | Sigma-Aldrich | Cat# G7021 |
| DL-Lactic Acid | Sigma-Aldrich | Cat# L4263 |
| BSA | MPBiomedicals | Cat# 810661 |
| L-Glutamine | Sigma-Aldrich | Cat# G7513 |
| 2-Mercaptoethanol | Sigma-Aldrich | Cat# M3148 |
| MEM Vitamin | Invitrogen | Cat# 11120–052 |
| NEAA | Invitrogen | Cat# 11140–050 |
| Ascorbic Acid | Sigma-Aldrich | Cat# A4544 |
| d-Biotin | Sigma-Aldrich | Cat# B4501 |
| Beta-Estradiol | Sigma-Aldrich | Cat# E2758 |
| FBS | Hyclone | Cat# SH30396.03 |
| KSR | Invitrogen | Cat# 10828–028 |
| Human FGF2 | Peprotech | Cat# 100–18B |
| Rat GDNF | Peprotech | Cat# 450–51 |
| Critical commercial assays | ||
| Slc7a5 probe | RNAscope® Probe | Cat# 461031 |
| Slc38a2 probe | RNAscope® Probe | Cat# 1184001-C2 |
| Slc2a1 probe | RNAscope® Probe | Cat# 458671-C2 |
| TruSeq RNA Sample Preparation Kit v2 | Illumina | Cat# RS-122–2001 |
| Illumina ChIP-Seq Sample Prep Kit | Illumina | Cat# FC-102–1003 |
| Illumina Sequencing Kit v4 | Illumina | Cat# 15003926 |
| Illumina TruSeq ChIP Sample Preparation Kit | Illumina | Cat# IP-202–1012 |
| Chromium Single Cell 3′ v3 Reagent Kits & Chip Kits | 10x Genomics | Cat# PN-1000075 |
| Seahorse XF Mito Fuel Flex | Agilent | Cat# 103260–100 |
| Glutamine/glutamate-Glo Assay kit | Promega | J8021 |
| NovaRed Kit | Vector Laboratories | SK-4800 |
| DC protein assay | BioRad | Cat# 5000111 |
| Deposited Data | ||
| RNA-Seq of WT, Meiosin KO and Stra8 KO Testes | Ishiguro et al.12 | DDBJ DRA007066 |
| ChIP-Seq of MEIOSIN | Ishiguro et al.12 | DDBJ DRA007778 |
| RNA-Seq and ChIP-Seq of GS Cells | Ishiguro et al.12 | GEO GSE116798 |
| STRA8-FLAG ChIP-Seq Data | Kojima et al.11 | GEO GSE115928 |
| Raw and analyzed 10x Genomics scRNA-seq datasets | This paper | GEO GSE211390 |
| In vitro meiosis 10x Genomics scRNA-seq | Zhang et al.15 | GEO GSE153300 |
| Mouse testis Slide-seq | Chen et al.26 | PRJNA668433 |
| Experimental models: Cell lines | ||
| Cultured mouse spermatogonial stem cells | This paper | N/A |
| Experimental models: organisms/strains | ||
| Mouse: C57BL/6J | Charles River Laboratory | Strain code: 027 |
| Mouse: Stra8Knockout | The Jackson Laboratory | JAX:023805 |
| MEF | R&D systems | Cat# PSC001 |
| Oligonucleotides | ||
| Slc7a5_siRNA | Dharmacon | Cat# 041166-01-0005 |
| Slc38a2 siRNA | Dharmacon | Cat# 059380-00-0005 |
| Slc2a1_siRNA | Dharmacon | Cat# 044254-02-0005 |
| Ctrl siRNA | Dharmacon | Cat# D-001210-01-05 |
| Primers for qRT-PCR | This paper | Table S1 |
| Recombinant DNA | ||
| Stra8-flag | This paper | N/A |
| Stra8Δ1 | This paper | N/A |
| Stra8Δ2 | This paper | N/A |
| Stra8Δ3 | This paper | N/A |
| Stra8ΔC | This paper | N/A |
| Software and algorithms | ||
| FlowJo v10 | FlowJo, LLC | https://www.flowjo.com/ |
| Cell Ranger v5.1.0 | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest |
| Seurat R package Version 3.0 | Butler et al.42 | https://satijalab.org/seurat/ |
| R Version 3.4 or higher | N/A | https://www.r-project.org |
| R studio Version 1.0.143 | N/A | https://www.rstudio.com |
| GraphPad Prism (v9.0) | N/A | https://www.graphpad.com/scientific-software/prism/ |
| ImageJ v1.41 | NIH | https://imagej.net/ij/ |
| DropletUtils | Griffiths et al.43 | https://www.nature.com/articles/s41467-018-05083-x |
| DoubletFinder | McGinnis et al.44 | https://www.sciencedirect.com/science/article/pii/S2405471219300730 |
| Harmony | Korsunsky et al.45 | https://www.nature.com/articles/s41592-019-0619-0 |
| Pheatmap | Kolde.46 | https://cran.r-project.org/web/packages/pheatmap/index.html |
| ClusterProfiler | Yu et al.47 | https://www.liebertpub.com/doi/10.1089/omi.2011.0118?url_ver=Z39.88-2003&rfr_id=ori%3Arid%3Acrossref.org&rfr_dat=cr_pub++0pubmed& |
| AlphaFold2 | Jumper et al.48 | https://www.nature.com/articles/s41586-021-03819-2 |
| MMseqs2 | Steinegger et al.49 | https://www.nature.com/articles/nbt.3988 |
| PyMOL | DeLano et al.50 | https://pymol.org/2/ |
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Animals
All experiments involving animals were conducted according to the approved protocol by the Institutional Animal Care and Use Committee (IACUC) at the University of Kansas Medical Center in strict accordance with its regulatory and ethical guidelines. All animals were housed in a specified pathogen-free facility with a 12 h light/dark cycle. All animals had access to food and water ad libitum. Male mice at postnatal day 12 or 21 were used for the performed experiments in this study. Stra8-deficient mice in C57Bl/6 background were obtained from Jackson Laboratory (Jax stock number: 023805).
Human testis tissues
Human testis samples were obtained from organ donors (20–29 years) through the Center for Organ Recovery and Education with approval of the University of Pittsburgh Committee for Oversight of Research and Clinical Training Involving Decedents (CORID 686). Upon surgical removal, testis tissues were fixed by 4% paraformaldehyde overnight at 4°C. Fixed samples were then washed with cold PBS three times, embedded in paraffin, and sectioned at 5 μm.
Mouse spermatogonial stem cell (SSC) culture
We generated mouse SSC culture in both wild-type and Stra8-deficient backgrounds as previously reported.15 Briefly, to establish SSC culture, juvenile mouse testes from P6 pups were dissociated by a two-step enzymatic digestion procedure. After removing tunica albuginea, testes were minced into small pieces and transferred to a 15 mL tube containing 4 mL of 1 mg/mL collagenase IV (Thermo Fisher, Cat# 17104019) solution in HBSS. Tissues were incubated in 37 °C water bath for 20 min until tubules were dissociated. After washing with HBSS and collecting by centrifugation, the tubule fragments were incubated in 4 mL of 0.05% trypsin-EDTA solution in 37 °C water bath for another 5 min. Testicular cells were cultured on 0.1% gelatin-coated plates with complete SSC medium overnight. Then, differential plating was applied to enrich stem cells: non-adherent germ cells were collected, and adherent somatic cells, e.g., Sertoli cells, were discarded. Collected germ cells were transfer on to irradiated mouse embryonic fibroblasts (MEF) (R&D system, Cat# PSC001) as feeder cells for expansion and long-term culture.
METHOD DETAILS
RNA isolation and qPCR
Total RNA was isolated from collected tissues or cells by using TRIzol™ (Invitrogen,15596018). The RT reaction was carried out with SuperScript II First-Strand Synthesis Kit (Invitrogen,18080–051). Quantitative polymerase chain reactions (qPCRs) were performed by using gene-specific primers (Table S1) and Power SYBR Green Master Mix (Applied Biosystems) on Applied Biosystems Quant Studio 5.
Immunocytofluorescence (ICF) analysis
For ICF staining, cultured cells were fixed with 4% paraformaldehyde (PFA) containing 0.1% Triton X-100 for 10 min at room temperature. Then cells were washed with PBS and blocked with 0.1% Triton X-100 and 5% BSA for 1 h at room temperature. The primary antibodies (Table S1) were added and incubated for overnight at 4 °C. After washing using PBS, the secondary antibodies were added and incubated for another 1 h at room temperature. Images were captured using Nikon A1R confocal microscope and were processed using Nikon NIS Elements and Adobe Photoshop softwares. To quantify signal intensity, we used the protocol from: https://www.unige.ch/medecine/bioimaging/files/1914/1208/6000/Quantification.pdf.
Immunohistochemistry (IHC)
For IHC staining, sections were treated with xylene for deparaffinization and ethanol (95%, 80%, and 75%) for rehydration. Antigen retrieval was achieved by microwaving in 10 mM citrate-based buffer at pH 6.0. Sections were washed by PBS with 0.05% Tween 20, followed by incubation with primary antibodies overnight at 4°C and secondary antibodies at room temperature for 1 h. The bound antibodies were detected by using NovaRed Kit (SK-4800, Vector Laboratories). Representative images were captured by using a NIS-Elements AR v5.10 (Nikon) and processed by NIS-Elements Viewer v5.21 (Nikon).
siRNA knockdown
For all siRNA knockdown experiments, cells were seeded at the same density and transfected on the following day with ON-TARGET plus single siRNAs or SMART pool siRNAs (a mixture of four siRNAs provided as a single reagent) (Dharmacon) by Lipofectamine 3000 (Invitrogen, L3000015) according to the manufacturer’s instructions. The sequences of the siRNAs are displayed in Table S1.
Western blot analysis
Total protein was isolated in RIPA buffer contained with 1 mM PMSF (Sigma) and protease inhibitor cocktail (Sigma P8340). Cell debris was removed by centrifugation at 12,000 × g for 15 min at 4 °C, and protein concentrations in supernatants were determined by DC protein assay (BioRad). Equal amount of protein from each sample was mixed with 4XLDS sample buffer (Invitrogen) plus 10X sample reducing agent (Invitrogen) and denatured for 10 min at 70 °C. Proteins were resolved in 4–12% Bis-Tris gels (Thermo Fisher) and transferred to PVDF membranes. Blots were probed with antibodies overnight at 4 °C, washed and reacted with secondary antibody. Detection was performed with the Clarity ECL Western Blotting Substrate (BioRad).
Single-cell RNA-seq (scRNA-seq)
The 10X Genomics Single-Cell 3′ Expression library preparation is performed using the 10X Genomics Chromium Controller. The cells prepared from disassociated tissue or tissue culture are validated for viability and cell concentration using the Countess II FL Automated Cell Counter (Life Technologies) targeting ≥75% cell viability. If debris or cell clumping is present in the cell suspension, the preparation is filtered through an FLOWMI Cell Strainer, 40 μm (Thermo Fisher 50-136-7555) to yield a homogeneous single-cell suspension. Cell counts are redetermined by using the Countess ll FL and adjusted to ~1000 cells/μl by low speed centrifugation at 4 °C and resuspended in 1×PBS without calcium or magnesium (Thermo Fisher MT21040CV) supplemented with 0.04% BSA to prepare cells for emulsification. The cell emulsification is performed with the 10× Chromium Controller using the Chromium Next GEM Single-Cell 3′ GEM Library & Gel Bead Kit v3.1 (10X Genomics 1000120) and Chromium Next GEM Chip G Single-Cell Kit (10X Genomics 1000127). Well 1 of Chip G is loaded with the RT Master Mix + cell suspension containing ~16,000 cells to target 10,000 emulsified cells at ~65% efficiency of emulsification. Well 2 of Chip G is filled with 50 μL of the Next GEM GEL Beads. Well 3 of Chip G is filled with 45 μL Partitioning Oil. Any unused wells are filled with 50% glycerol at a volume designated for the well number. A gasket is applied to the Chip G and the loading cassette and inserted into the Chromium Controller for GEM creation Using the Chromium Single-Cell G run program. Emulsified GEMs are recovered from each well and transferred to 200 μL strip tubes. The RT reaction to generate 10X barcoded single stranded cDNA, in the single-cell containing GEMs, is performed using an Eppendorf MasterCycler Pro thermal cycler. Post GEM-RT Cleanup is conducted by breaking the GEMS with 10X Recovery Agent to separate the aqueous phase from the Recovery Agent and Partitioning oil. A cleanup of the sscDNA containing aqueous phase is completed using Dynabeads MyOne Silane beads (Life Technologies 37002D). The second strand cDNA amplification is performed using the cDNA Amplification Mix on the Eppendorf MasterCycler Pro. The 3′ gene expression library construction is initiated with fragmentation, end repair and A-tailing of the dscDNA followed by adapter ligation and a sample index PCR using the Illumina compatible indexed adapters in the Chromium i7 Multiplex kit (10X Genomics 120262). Validation of the single-cell library is conducted using the Agilent Tapestation 4200 ScreenTape assay (Agilent 5067–5576). Single-cell library quantification is completed using a Roche LightCycle96 using FastStart Essential Green Master (Roche 06402712001) and KAPA Library Quant (Illumina) DNA Standards (KAPA KK4903). Library concentrations are adjusted to 3 nm and pooled. The library pool is diluted to 1.00 nm for a final clustering concentration of 200pM on a NovaSeq6000. The sequencing was performed using a NovaSeq6000 100 cycle Reagent Kit (Illumina 20012865) with an asymmetrical sequencing profile (read 1–28 cycle: i7 index read—8 cycle: i5 index read—0 cycle: read 2–94 cycles). Bcl2fastq conversion and demultiplexing is performed using the 10X Genomics Cell Ranger and Loupe Browser software suite.
Analysis of single-cell RNA-seq data
Illumina base call files for all libraries were demultiplexed and converted to FASTQ files using bcl2fastq v.2.20.0.422 (Illumina). The CellRanger pipeline (10X Genomics, v.5.1.0) was used to align reads to the human reference mm10, deduplicate reads, call cells and generate cell-by-gene digital counts matrices for each library. The resultant count matrices were further processed with Seurat (https://satijalab.org/seurat/, R package, v3.1)42 to exclude genes that were detected in less than 3 cells and to exclude cells with the following parameters: less than 1,000 unique molecular identifiers (UMIs), maximum mitochondrial content of 5%. Doublet and droplet identification were performed using DropletUtils43,51 and DoubletFinder.44 Filtered matrices were then combined and normalized such that the number of UMIs in each cell was equal to the median UMI across the dataset and log-transformed. Seurat was used to identify the top 2,000 highly variable genes from the log-transformed combined matrix. The mitochondrial genes, hemoglobin genes and ribosomal genes were excluded from the highly variable gene set. Principal component analysis and neighborhood graph generation were performed based on the highly variable genes set. Harmony (v.1.0) batch correction45 was performed to reduce variabilities introduced by inherent inter-individual differences. Batched-corrected principal components were used for dimensionality reduction using uniform manifold approximation and projection (UMAP). Pheatmap package was used for heatmap plotting and clustering.46 GO analysis of genes were analyzed by using the ClusterProfiler package.47 DEGs were generated by code published with modifications.52
Human STRA8 and MEIOSIN complexes structural models were predicted by AlphaFold250, which supports protein structure prediction using MSAs generated by MMseqs251. PyMOL50 is used for generating 3D structure.
Seahorse mito fuel flex
Mito fuel flex tests were performed with Seahorse kits and the Seahorse XFe96 analyzer according to the manufacturer’s protocol (Agilent). In brief, F9 embryonic carcinoma cells were seeded in a Seahorse XF96 cell culture plate and incubated at 37 °C in a humidified 5% CO2 atmosphere overnight. Seahorse XF cartridges were hydrated with Seahorse XF calibrant and placed in a 0% CO2 incubator overnight. Prior to the assay, the culture medium was replaced with assay medium per the manufacturer’s instructions. Compound injections for different stress tests were prepared according to the manufacturer’s instructions (Seahorse XF Mito Fuel Flex Test Kit) and added to the respective ports on the cartridge. Three to five measurements were made for basal metabolism and each compound injection. Seahorse XF Report Generator (Agilent) was used to automatically calculates the key parameters of the Seahorse XF Mito Fuel Flex results per the manufacturer’s instruction.
Glutamine/glutamate-Glo assay
The concentration of intracellular glutamine and glutamate was measured in cell lysates using the glutamine/glutamate-Glo Assay kit (Promega J8021) following producer’s instructions. Luminescence was measured using an iD5 Plate Reader.
FACS and glucose-uptake assay
Single-cell suspensions from cultured SSCs or split from testis mechanical separation through a 70-μm filter. Single-cell suspensions from cultured SSCs or F9 cells were placed in serum-free DMEM containing 40 μM 2-NBDG or 0.4 μM glucose-Cy5 for 25 min at 37 °C. Glucose-Cy5, a Cy5-linked 1-amino-glucose tracer, was bought from Sigma (Cat# SML3233). Analysis of SSC markers (a6-integrin and Kit) was performed using the BD according to manufacturer’s protocol. 1 × 106 cells were incubated in primary antibody at the recommended concentrations for 1 h on ice and protected from light. Cells were subsequently washed in PBS and counterstained with DAPI to exclude dead cells. Flow cytometric analysis and sorting was performed on a BD FACS Aria III.
QUANTIFICATION AND STATISTICAL ANALYSIS
All experiments were replicated at least three times independently. Different mice, tissues or cells were used during each experimental replicate. Quantitative data from the experimental replicates were pooled and are presented as mean ± SE as indicated in the figure legend. Compiled data were analyzed by Student’s t test and one-way ANOVA with Bonferroni’s multiple comparisons test.
Supplementary Material
Highlights.
Mouse germ cells withdraw nutrient transporter expression during meiotic initiation
This process is mediated by retinoic acid via its germline target Stra8
Knockdown of Slc38a2, a glutamine importer, inhibits mTORC1/PKA activities
Slc38a2 knockdown or mTORC1/PKA inhibition activates meiotic gene expression
ACKNOWLEDGMENTS
We thank Jing Huang at Kansas IDDRC Histology Services for sample processing and histological sections. We thank Xiaowan Wang and Heather Wilkins for help with the use of the Seahorse FX analyzer (supported by the University of Kansas Alzheimer’s Disease Research Center, P30AG035982). We thank Clark Bloomer, Rosanne Skinner, Veronica Cloud, and Yafen Niu at the University of Kansas Medical Center Genomics Core, which is supported by Kansas Intellectual and Developmental Disabilities Research Center (NIH U54 HD 090216), the Molecular Regulation of Cell Development and Differentiation—COBRE (P30 GM122731-03)—the NIH S10 High-End Instrumentation Grant (NIH S10OD021743), and the Frontiers CTSA grant (UL1TR002366). This work was supported by the National Institutes of Health (NIH) grants R01HD-103888, 3R01HD-103888-03, KUMC Research Institute, and the Department of Cell Biology and Physiology to N.W. X.Z. is a recipient of Kansas IDeA Network of Biomedical Research Excellence (K-INBRE) Developmental Research Project Program Award from the National Institute of General Medical Sciences of the NIH under grant number P20 GM103418. The content is solely the responsibility of the authors and does not necessarily represent the official views of the aforementioned funders.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112749.
DECLARATION OF INTERESTS
P.A.C. has served as a consultant for Pfizer Inc., Abbott Laboratories, and Janssen Research & Development.
REFERENCES
- 1.Handel MA, and Schimenti JC (2010). Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat. Rev. Genet 11, 124–136. 10.1038/nrg2723. [DOI] [PubMed] [Google Scholar]
- 2.Weidberg H, Moretto F, Spedale G, Amon A, and van Werven FJ (2016). Nutrient Control of Yeast Gametogenesis Is Mediated by TORC1, PKA and Energy Availability. PLoS Genet 12, e1006075. 10.1371/journal.pgen.1006075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tam J, and van Werven FJ (2020). Regulated repression governs the cell fate promoter controlling yeast meiosis. Nat. Commun 11, 2271. 10.1038/s41467-020-16107-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Werven FJ, and Amon A (2011). Regulation of entry into gametogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci 366, 3521–3531. 10.1098/rstb.2011.0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Rooij DG, and Russell LD (2000). All you wanted to know about spermatogonia but were afraid to ask. J. Androl 21, 776–798. [PubMed] [Google Scholar]
- 6.Nakagawa T, Sharma M, Nabeshima Y.i., Braun RE, and Yoshida S (2010). Functional hierarchy and reversibility within the murine spermatogenic stem cell compartment. Science 328, 62–67. 10.1126/science.1182868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith BE, and Braun RE (2012). Germ cell migration across Sertoli cell tight junctions. Science 338, 798–802. 10.1126/science.1219969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowles J, and Koopman P (2007). Retinoic acid, meiosis and germ cell fate in mammals. Development 134, 3401–3411. 10.1242/dev.001107. [DOI] [PubMed] [Google Scholar]
- 9.Baltus AE, Menke DB, Hu YC, Goodheart ML, Carpenter AE, de Rooij DG, and Page DC (2006). In germ cells of mouse embryonic ovaries, the decision to enter meiosis precedes premeiotic DNA replication. Nat. Genet 38, 1430–1434. 10.1038/ng1919. [DOI] [PubMed] [Google Scholar]
- 10.Anderson EL, Baltus AE, Roepers-Gajadien HL, Hassold TJ, de Rooij DG, van Pelt AM, and Page DC (2008). Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice. Proc. Natl. Acad. Sci. USA 105, 14976–14980. 10.1073/pnas.0807297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kojima ML, de Rooij DG, and Page DC (2019). Amplification of a broad transcriptional program by a common factor triggers the meiotic cell cycle in mice. Elife 8, e43738. 10.7554/eLife.43738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishiguro KI, Matsuura K, Tani N, Takeda N, Usuki S, Yamane M, Sugimoto M, Fujimura S, Hosokawa M, Chuma S, et al. (2020). MEIOSIN Directs the Switch from Mitosis to Meiosis in Mammalian Germ Cells. Dev. Cell 52, 429–445.e10. 10.1016/j.devcel.2020.01.010. [DOI] [PubMed] [Google Scholar]
- 13.Ferder IC, Fung L, Ohguchi Y, Zhang X, Lassen KG, Capen D, Brown D, Xavier RJ, and Wang N (2019). Meiotic gatekeeper STRA8 suppresses autophagy by repressing Nr1d1 expression during spermatogenesis in mice. PLoS Genet 15, e1008084. 10.1371/journal.pgen.1008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Efeyan A, Comb WC, and Sabatini DM (2015). Nutrient-sensing mechanisms and pathways. Nature 517, 302–310. 10.1038/nature14190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, Gunewardena S, and Wang N (2021). Nutrient restriction synergizes with retinoic acid to induce mammalian meiotic initiation in vitro. Nat. Commun 12, 1758. 10.1038/s41467-021-22021-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiong M, Ferder IC, Ohguchi Y, and Wang N (2015). Quantitative analysis of male germline stem cell differentiation reveals a role for the p53-mTORC1 pathway in spermatogonial maintenance. Cell Cycle 14, 2905–2913. 10.1080/15384101.2015.1069928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Q, Nie R, Li Y, Friel P, Mitchell D, Hess RA, Small C, and Griswold MD (2008). Expression of stimulated by retinoic acid gene 8 (Stra8) in spermatogenic cells induced by retinoic acid: an in vivo study in vitamin A-sufficient postnatal murine testes. Biol. Reprod 79, 35–42. 10.1095/biolreprod.107.066795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Endo T, Romer KA, Anderson EL, Baltus AE, de Rooij DG, and Page DC (2015). Periodic retinoic acid-STRA8 signaling intersects with periodic germ-cell competencies to regulate spermatogenesis. Proc. Natl. Acad. Sci. USA 112, E2347–E2356. 10.1073/pnas.1505683112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Song W, Wang J, Wang T, Xiong X, Qi Z, Fu W, Yang X, and Chen YG (2020). Single-cell transcriptome analysis reveals differential nutrient absorption functions in human intestine. J. Exp. Med 217, e20191130. 10.1084/jem.20191130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirsch D, Stahl A, and Lodish HF (1998). A family of fatty acid transporters conserved from mycobacterium to man. Proc. Natl. Acad. Sci. USA 95, 8625–8629. 10.1073/pnas.95.15.8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bröer A, Rahimi F, and Bröer S (2016). Deletion of Amino Acid Transporter ASCT2 (SLC1A5) Reveals an Essential Role for Transporters SNAT1 (SLC38A1) and SNAT2 (SLC38A2) to Sustain Glutaminolysis in Cancer Cells. J. Biol. Chem 291, 13194–13205. 10.1074/jbc.M115.700534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanai Y, Segawa H, Miyamoto K.i., Uchino H, Takeda E, and Endou H (1998). Expression cloning and characterization of a transporter for large neutral amino acids activated by the heavy chain of 4F2 antigen (CD98). J. Biol. Chem 273, 23629–23632. 10.1074/jbc.273.37.23629. [DOI] [PubMed] [Google Scholar]
- 23.Mueckler M, Caruso C, Baldwin SA, Panico M, Blench I, Morris HR, Allard WJ, Lienhard GE, and Lodish HF (1985). Sequence and structure of a human glucose transporter. Science 229, 941–945. 10.1126/science.3839598. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Pavlova NN, and Thompson CB (2017). Cancer cell metabolism: the essential role of the nonessential amino acid, glutamine. EMBO J 36, 1302–1315. 10.15252/embj.201696151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soumillon M, Necsulea A, Weier M, Brawand D, Zhang X, Gu H, Barthès P, Kokkinaki M, Nef S, Gnirke A, et al. (2013). Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep 3, 2179–2190. 10.1016/j.celrep.2013.05.031. [DOI] [PubMed] [Google Scholar]
- 26.Chen H, Murray E, Sinha A, Laumas A, Li J, Lesman D, Nie X, Hotaling J, Guo J, Cairns BR, et al. (2021). Dissecting mammalian spermatogenesis using spatial transcriptomics. Cell Rep 37, 109915. 10.1016/j.celrep.2021.109915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oulad-Abdelghani M, Bouillet P, Décimo D, Gansmuller A, Hey-berger S, Dollé P, Bronner S, Lutz Y, and Chambon P (1996). Characterization of a premeiotic germ cell-specific cytoplasmic protein encoded by Stra8, a novel retinoic acid-responsive gene. J. Cell Biol 135, 469–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koubova J, Menke DB, Zhou Q, Capel B, Griswold MD, and Page DC (2006). Retinoic acid regulates sex-specific timing of meiotic initiation in mice. Proc. Natl. Acad. Sci. USA 103, 2474–2479. 10.1073/pnas.0510813103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Velte EK, Niedenberger BA, Serra ND, Singh A, Roa-DeLaCruz L, Hermann BP, and Geyer CB (2019). Differential RA responsiveness directs formation of functionally distinct spermatogonial populations at the initiation of spermatogenesis in the mouse. Development 146, dev173088. 10.1242/dev.173088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. (2010). Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 107, 21931–21936. 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park S, Kim GW, Kwon SH, and Lee JS (2020). Broad domains of histone H3 lysine 4 trimethylation in transcriptional regulation and disease. FEBS J 287, 2891–2902. 10.1111/febs.15219. [DOI] [PubMed] [Google Scholar]
- 32.GTEx Consortium (2013). The Genotype-Tissue Expression (GTEx) project. Nat. Genet 45, 580–585. 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leu JY, Chua PR, and Roeder GS (1998). The meiosis-specific Hop2 protein of S. cerevisiae ensures synapsis between homologous chromosomes. Cell 94, 375–386. 10.1016/s0092-8674(00)81480-4. [DOI] [PubMed] [Google Scholar]
- 34.Petukhova GV, Romanienko PJ, and Camerini-Otero RD (2003). The Hop2 protein has a direct role in promoting interhomolog interactions during mouse meiosis. Dev. Cell 5, 927–936. 10.1016/s1534-5807(03)00369-1. [DOI] [PubMed] [Google Scholar]
- 35.Mark M, Jacobs H, Oulad-Abdelghani M, Dennefeld C, Féret B, Vernet N, Codreanu CA, Chambon P, and Ghyselinck NB (2008). STRA8-deficient spermatocytes initiate, but fail to complete, meiosis and undergo premature chromosome condensation. J. Cell Sci 121, 3233–3242. 10.1242/jcs.035071. [DOI] [PubMed] [Google Scholar]
- 36.Tang JX, Li J, Cheng JM, Hu B, Sun TC, Li XY, Batool A, Wang ZP, Wang XX, Deng SL, et al. (2017). Requirement for CCNB1 in mouse spermatogenesis. Cell Death Dis 8, e3142. 10.1038/cddis.2017.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen T, Sun Y, Ji P, Kopetz S, and Zhang W (2015). Topoisomerase IIα in chromosome instability and personalized cancer therapy. Oncogene 34, 4019–4031. 10.1038/onc.2014.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leduc F, Maquennehan V, Nkoma GB, and Boissonneault G (2008). DNA damage response during chromatin remodeling in elongating spermatids of mice. Biol. Reprod 78, 324–332. 10.1095/biolreprod.107.064162. [DOI] [PubMed] [Google Scholar]
- 39.Bergström J, Fürst P, Norée LO, and Vinnars E (1974). Intracellular free amino acid concentration in human muscle tissue. J. Appl. Physiol 36, 693–697. 10.1152/jappl.1974.36.6.693. [DOI] [PubMed] [Google Scholar]
- 40.Cheng CY, and Mruk DD (2012). The blood-testis barrier and its implications for male contraception. Pharmacol. Rev 64, 16–64. 10.1124/pr.110.002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mruk DD, and Cheng CY (2015). The Mammalian Blood-Testis Barrier: Its Biology and Regulation. Endocr. Rev 36, 564–591. 10.1210/er.2014-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Griffiths JA, Richard AC, Bach K, Lun ATL, and Marioni JC (2018). Detection and removal of barcode swapping in single-cell RNA-seq data. Nat. Commun 9, 2667. 10.1038/s41467-018-05083-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McGinnis CS, Murrow LM, and Gartner ZJ (2019). DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst 8, 329–337.e4. 10.1016/j.cels.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh PR, and Raychaudhuri S (2019). Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296. 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kolde R (2012). Package ‘pheatmap’. Bioconductor 1–6. [Google Scholar]
- 47.Yu G, Wang LG, Han Y, and He QY (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steinegger M, and Söding J (2017). MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol 35, 1026–1028. 10.1038/nbt.3988. [DOI] [PubMed] [Google Scholar]
- 50.DeLano WL (2002). Pymol: An open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr 40, 82–92. [Google Scholar]
- 51.Lun ATL, Riesenfeld S, Andrews T, Dao TP, Gomes T, and participants in the 1st Human Cell Atlas Jamboree; and Marioni JC (2019). EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol 20, 63. 10.1186/s13059-019-1662-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stévant I, Neirijnck Y, Borel C, Escoffier J, Smith LB, Antonarakis SE, Dermitzakis ET, and Nef S (2018). Deciphering Cell Lineage Spec-ification during Male Sex Determination with Single-Cell RNA Sequencing. Cell Rep 22, 1589–1599. 10.1016/j.celrep.2018.01.043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
FastQ files of single-cell RNA-seq data have been deposited at the Gene Expression Omnibus (GEO) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PLZF | Santa Cruz | Cat# sc-28319; RRID:AB_2218941 |
| KIT | R&D Systems | Cat# AF1356; RRID:AB_354750 |
| SLC7A5 | Cell Signaling Technology | Cat# 5347S; RRID:AB_10695104 |
| SLC38A2 | Sigma Aldrich | Cat# HPA035180; RRID:AB_10669901 |
| SLC2A1 | Abcam | Cat# ab115730; RRID:AB_10903230 |
| gH2AX | Millpore | Cat# 05–636; RRID:AB_309864 |
| VASA | R&D Systems | Cat# AF2030; RRID:AB_2277369 |
| P70S6K | Cell Signaling Technology | Cat# 9234P; RRID:AB_2269803 |
| p-70S6K | Cell Signaling Technology | Cat# 9202S; RRID:AB_331676 |
| S6 | Cell Signaling Technology | Cat# 4858P; RRID:AB_916156 |
| p-S6 | Cell Signaling Technology | Cat# 2217S; RRID:AB_331355 |
| 4EBP1 | Cell Signaling Technology | Cat# 2855P; RRID:AB_560835 |
| p-4EBP1 | Cell Signaling Technology | Cat# 9644T; RRID:AB_2097841 |
| FLAG | Sigma Aldrich | Cat# F1804; RRID:AB_262044 |
| a6 integrin | BD Biosciences | Cat# 555736; RRID:AB_396079 |
| KIT | BD Biosciences | Cat# 553356; RRID:AB_398536 |
| donkey anti-mouse IgG Alexa Flour 488 | Life Technologies | Cat# A21202; RRID:AB_141607 |
| donkey anti-rabbit IgG Alexa Flour 568 | Life Technologies | Cat# A10042; RRID:AB_2534017 |
| donkey anti-goat IgG Alexa Flour 647 | Life Technologies | Cat# A21447; RRID:AB_141844 |
| Biological samples | ||
| Human testis testicular tissue from organ donors | Center for Organ Recovery and Education | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Superscript II Reverse Transcriptase | Thermo Fisher Scientific | Cat# 18080051 |
| NuPage 4 %–12 %Bis-Tris Protein Gel | Thermo Fisher Scientific | Cat# NP0322BOX |
| cOmplete, EDTA-Free | Roche | Cat# 4 693 132 |
| RIPA Buffer | Thermo | Cat# 89900 |
| all-transRA | Sigma-Aldrich | Cat# R2625 |
| 2-NBDG | Sigma-Aldrich | Cat# 72987 |
| Glucose-Cy5 | Sigma-Aldrich | Cat# SML3233 |
| collagenase IV | Thermo Fisher Scientific | Cat# 17104019 |
| TRIzol | Invitrogen | Cat# 15596018 |
| Power SYBR Green Master Mix | Applied Biosystems | Cat# A25742 |
| Lipofectamine 3000 | Invitrogen | Cat# L3000015 |
| N2 | R&D Systems | Cat# AR-009 |
| Pyruvic Acid | Sigma-Aldrich | Cat# P2256 |
| D-(+)-glucose | Sigma-Aldrich | Cat# G7021 |
| DL-Lactic Acid | Sigma-Aldrich | Cat# L4263 |
| BSA | MPBiomedicals | Cat# 810661 |
| L-Glutamine | Sigma-Aldrich | Cat# G7513 |
| 2-Mercaptoethanol | Sigma-Aldrich | Cat# M3148 |
| MEM Vitamin | Invitrogen | Cat# 11120–052 |
| NEAA | Invitrogen | Cat# 11140–050 |
| Ascorbic Acid | Sigma-Aldrich | Cat# A4544 |
| d-Biotin | Sigma-Aldrich | Cat# B4501 |
| Beta-Estradiol | Sigma-Aldrich | Cat# E2758 |
| FBS | Hyclone | Cat# SH30396.03 |
| KSR | Invitrogen | Cat# 10828–028 |
| Human FGF2 | Peprotech | Cat# 100–18B |
| Rat GDNF | Peprotech | Cat# 450–51 |
| Critical commercial assays | ||
| Slc7a5 probe | RNAscope® Probe | Cat# 461031 |
| Slc38a2 probe | RNAscope® Probe | Cat# 1184001-C2 |
| Slc2a1 probe | RNAscope® Probe | Cat# 458671-C2 |
| TruSeq RNA Sample Preparation Kit v2 | Illumina | Cat# RS-122–2001 |
| Illumina ChIP-Seq Sample Prep Kit | Illumina | Cat# FC-102–1003 |
| Illumina Sequencing Kit v4 | Illumina | Cat# 15003926 |
| Illumina TruSeq ChIP Sample Preparation Kit | Illumina | Cat# IP-202–1012 |
| Chromium Single Cell 3′ v3 Reagent Kits & Chip Kits | 10x Genomics | Cat# PN-1000075 |
| Seahorse XF Mito Fuel Flex | Agilent | Cat# 103260–100 |
| Glutamine/glutamate-Glo Assay kit | Promega | J8021 |
| NovaRed Kit | Vector Laboratories | SK-4800 |
| DC protein assay | BioRad | Cat# 5000111 |
| Deposited Data | ||
| RNA-Seq of WT, Meiosin KO and Stra8 KO Testes | Ishiguro et al.12 | DDBJ DRA007066 |
| ChIP-Seq of MEIOSIN | Ishiguro et al.12 | DDBJ DRA007778 |
| RNA-Seq and ChIP-Seq of GS Cells | Ishiguro et al.12 | GEO GSE116798 |
| STRA8-FLAG ChIP-Seq Data | Kojima et al.11 | GEO GSE115928 |
| Raw and analyzed 10x Genomics scRNA-seq datasets | This paper | GEO GSE211390 |
| In vitro meiosis 10x Genomics scRNA-seq | Zhang et al.15 | GEO GSE153300 |
| Mouse testis Slide-seq | Chen et al.26 | PRJNA668433 |
| Experimental models: Cell lines | ||
| Cultured mouse spermatogonial stem cells | This paper | N/A |
| Experimental models: organisms/strains | ||
| Mouse: C57BL/6J | Charles River Laboratory | Strain code: 027 |
| Mouse: Stra8Knockout | The Jackson Laboratory | JAX:023805 |
| MEF | R&D systems | Cat# PSC001 |
| Oligonucleotides | ||
| Slc7a5_siRNA | Dharmacon | Cat# 041166-01-0005 |
| Slc38a2 siRNA | Dharmacon | Cat# 059380-00-0005 |
| Slc2a1_siRNA | Dharmacon | Cat# 044254-02-0005 |
| Ctrl siRNA | Dharmacon | Cat# D-001210-01-05 |
| Primers for qRT-PCR | This paper | Table S1 |
| Recombinant DNA | ||
| Stra8-flag | This paper | N/A |
| Stra8Δ1 | This paper | N/A |
| Stra8Δ2 | This paper | N/A |
| Stra8Δ3 | This paper | N/A |
| Stra8ΔC | This paper | N/A |
| Software and algorithms | ||
| FlowJo v10 | FlowJo, LLC | https://www.flowjo.com/ |
| Cell Ranger v5.1.0 | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest |
| Seurat R package Version 3.0 | Butler et al.42 | https://satijalab.org/seurat/ |
| R Version 3.4 or higher | N/A | https://www.r-project.org |
| R studio Version 1.0.143 | N/A | https://www.rstudio.com |
| GraphPad Prism (v9.0) | N/A | https://www.graphpad.com/scientific-software/prism/ |
| ImageJ v1.41 | NIH | https://imagej.net/ij/ |
| DropletUtils | Griffiths et al.43 | https://www.nature.com/articles/s41467-018-05083-x |
| DoubletFinder | McGinnis et al.44 | https://www.sciencedirect.com/science/article/pii/S2405471219300730 |
| Harmony | Korsunsky et al.45 | https://www.nature.com/articles/s41592-019-0619-0 |
| Pheatmap | Kolde.46 | https://cran.r-project.org/web/packages/pheatmap/index.html |
| ClusterProfiler | Yu et al.47 | https://www.liebertpub.com/doi/10.1089/omi.2011.0118?url_ver=Z39.88-2003&rfr_id=ori%3Arid%3Acrossref.org&rfr_dat=cr_pub++0pubmed& |
| AlphaFold2 | Jumper et al.48 | https://www.nature.com/articles/s41586-021-03819-2 |
| MMseqs2 | Steinegger et al.49 | https://www.nature.com/articles/nbt.3988 |
| PyMOL | DeLano et al.50 | https://pymol.org/2/ |


