Figure 3. BLADE SELEX generates cleavage capable sgRNA scaffolds.
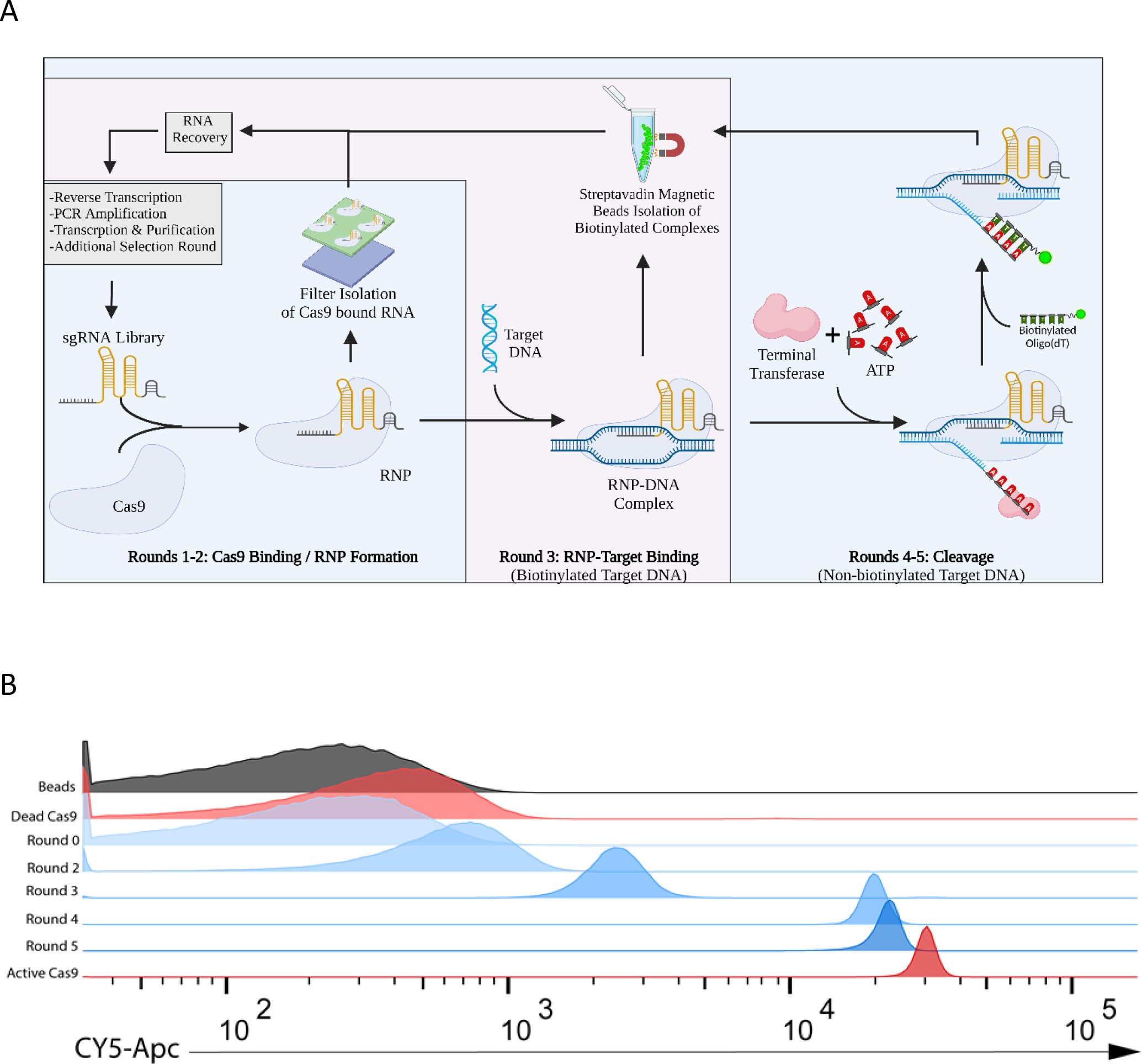
(A) A library based on the WT SpCas9 sgRNA sequence was synthesized with 5’ and 3’ constant regions and a 60 nucleotide partially randomized region. BLADE SELEX was performed with this library and consisted of 3 phases: RNA pool-RNP formation, RNP binding to DNA substrate, and a TdT-based screen for cleavage. (B) DNA cleavage by the pools of sgRNA variants isolated following various rounds of BLADE SELEX (blue) was assessed by A-tailing and flow cytometry and compared to the WT sgRNA (red) complexed with inactive “dead” Cas9 or active Cas9. In this assay, functional RNPs cleave a Cy5-labeled DNA target, TdT adds a poly(A) tail to the newly exposed 3’ end-cleavage product allowing for capture by a biotinylated oligo(dT) probe and magnetic streptavidin beads.
