Figure: Chromatin and transcriptomic shifts during renin cell development.
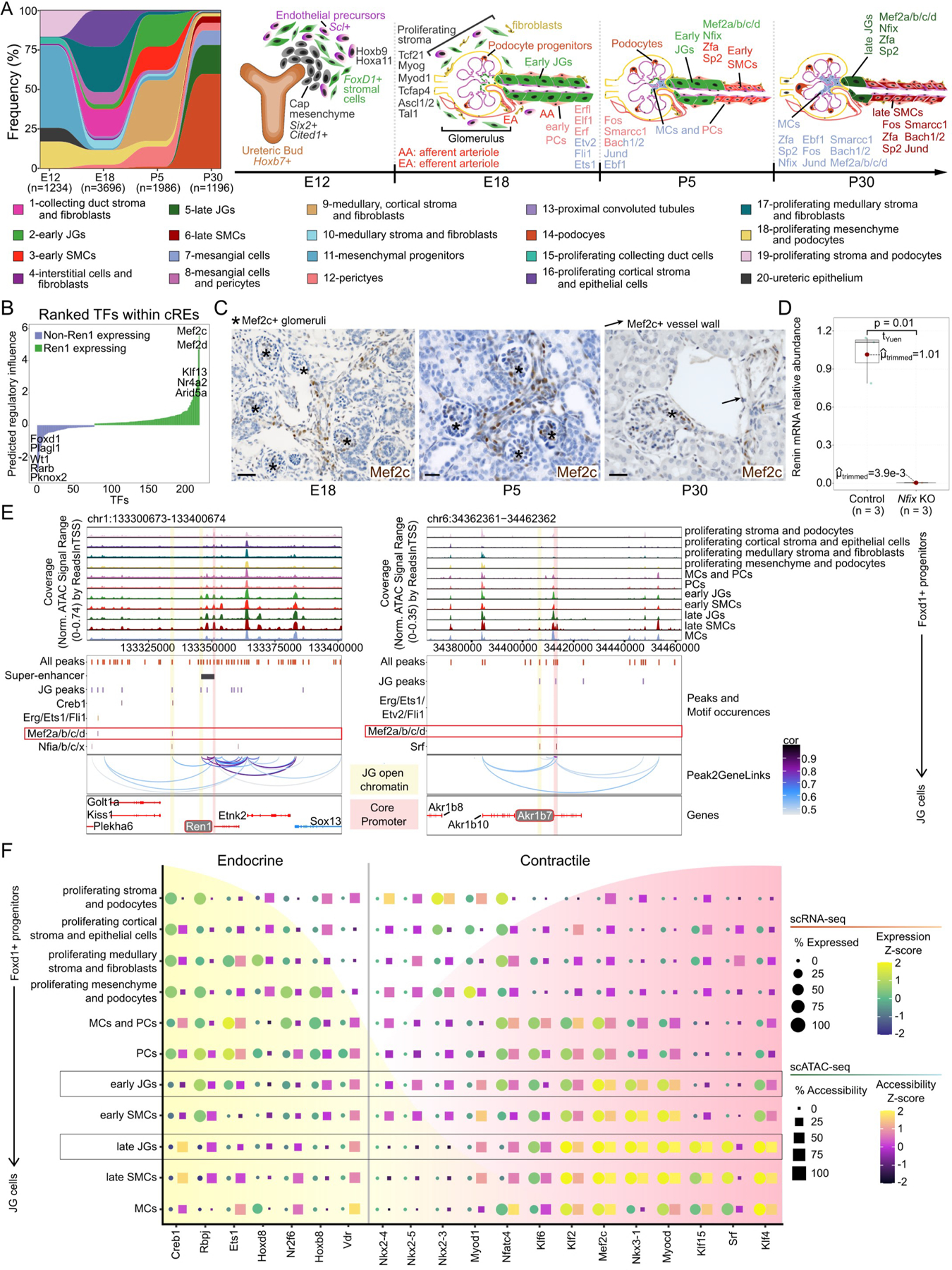
(A), Alluvial plot of cell frequency distribution (left) and individually enriched TF motifs (right) across developmental time points. Hoxb9 and Hoxa11 are enriched at E12. At E18, Tcf21, Myog, Myod1, Tcfap4, Ascl1 and Ascl2, and Tal1 are enriched in proliferating stroma. Between E18 and P5, Erfl, Elf1, Erf, Etv2, Fli1, Fos, Smarcc1, Bach1 and Bach2, Jund, and Ebf1 are enriched in early PCs and mixed MCs and PCs. In early JGs and SMCs, Mef2a/b/c/d, Nfix, Zfa, and Sp2 motifs are enriched. By P30, MCs are enriched for Fos, Smarcc1, Bach1 and Bach2, Jund, Ebf1, and to a lesser extent Mef2a/b/c/d, Nfix, Zfa, and Sp2. Late JGs are enriched for Mef2a/b/c/d, Nfix, Zfa, and Sp2 motifs, and late SMCs for Fos, Smarcc1, Bach1 and Bach2, Jund, Ebf1, Zfa, and Sp2. (B), TFs ranked by predicted regulatory score. (C), Mef2c IHC in kidney sections at E18, P5, and P30. At E18, Mef2c is found along AAs, interlobular arterioles, and MCs while at P5, it is found inside glomeruli, MCs, AAs, interlobular arterioles, and at the JGA. Scale bars: 50μm. (D), Deletion of Nfix in cultured As4.1 cells decreases Ren1 mRNA (n=3, p=0.01, Yuen-Welch’s test). Three gRNAs were designed for Nfix knockout with 92% transfection efficiency. (E), Browser tracks identify enriched open chromatin and TF motif sites at Ren1 (left) and Akr1b7 (right). (F), Dot plot for markers of endocrine or contractile phenotypes throughout JG cell differentiation.
