Figure 1. Replacement of histone H3.1 by H3.3 in ddm1 mutants.
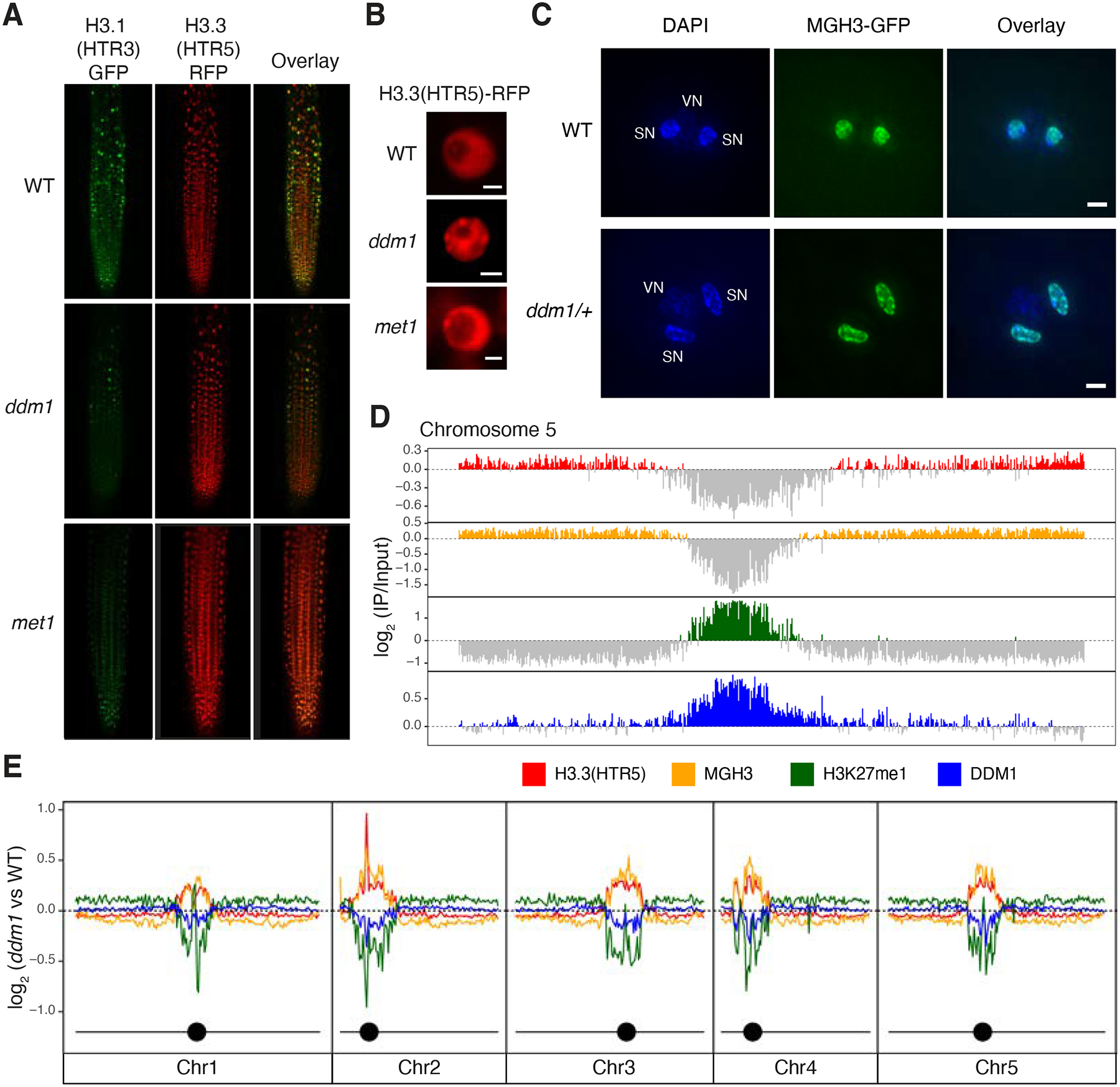
(A) H3.1(HTR3)-GFP and H3.3(HTR5)-RFP localization in Arabidopsis root tips of wild-type (WT), ddm1, and met1. (B) Ectopic chromocenter localization of H3.3(HTR5)-RFP in ddm1 as compared to WT and met1. Scale bars indicate 2 μm. (C) Male Germline-specific Histone H3.3 variant MGH3-GFP localization in sperm nuclei of Arabidopsis pollen grains from WT and ddm1/+ plants. DAPI staining was used to visualize vegetative (VN) and sperm nuclei (SN). Mis-localization to the nuclear periphery was observed in pollen from ddm1/+. Scale bars indicate 2 μm. (D) Distribution of ChIP-seq marks in WT along chromosome 5, showing preferential localization of H3.3(HTR5) in leaf tissue and MGH3 in pollen on chromosome arms, and H3K27me1 and DDM1 on pericentromeric heterochromatin. The values correspond to the log2 fold change of IP/H3 for H3.3(HTR5) and H3K27me1, and IP/Input for DDM1 and MGH3, normalized in counts per million. Signal tracks were averaged in 50kb windows with negative log2 values shown in grey. (E) Distribution of the log2 ratio of the ChIP-seq coverage between ddm1 and WT, showing an increase in H3.3 and MGH3 in peri-centromeric regions, coupled with a loss of H3K27me1 and DDM1. MGH3 IP was performed on pollen grains from a heterozygote ddm1/+ plant (as in C). In ddm1-2 mutants, DDM1 protein is present at reduced levels (Figure S1E).
