Figure 4. Structure and function of the Helicase C terminal and ATPase lobes.
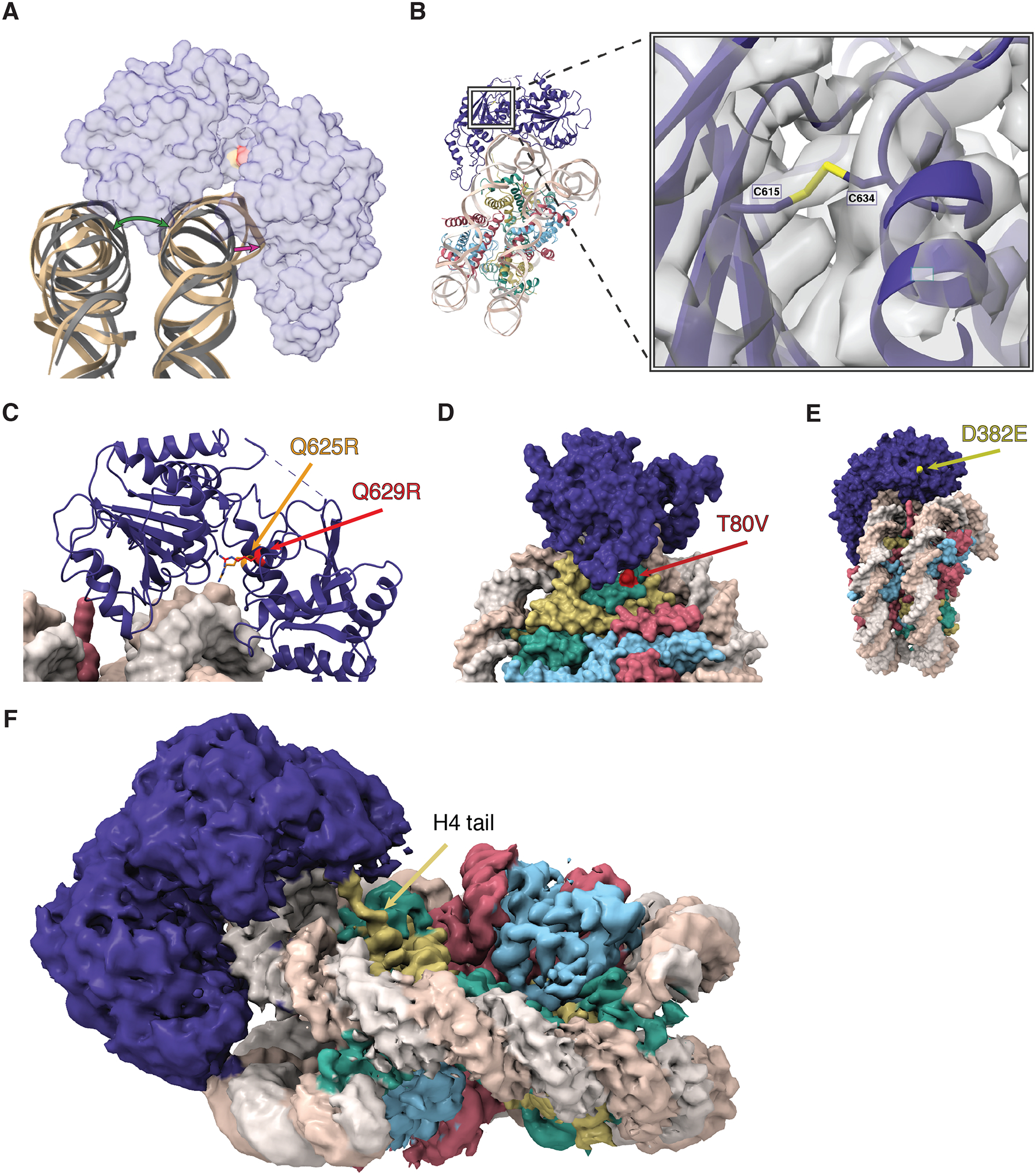
(A) A cartoon view of the DNA distortion caused by DDM1 binding. The DNA backbone of the DDM1-nucleosome model (tan) was aligned to a naked nucleosome DNA backbone (grey, PDB code 1KX5), showing the distortion of DNA where DDM1 is bound to the nucleosome, as well as distortion on the other gyre. A transparent surface model of DDM1 is shown for clarity. The green arrow shows the distortion (opening) of the gyre position caused by DDM1 binding to the nucleosome, and the magenta arrow represents the distortion of the DNA backbone. (B) A view of the disulfide bond formed between C615 and C634, connecting two regions in the HELICc domain of DDM1. The molecular model is shown as ribbons, cryo-EM density is shown as a gray volume. The first mutation of ddm1 to be isolated, ddm1-1, substitutes C615 for Y and has a strong DNA methylation defect. (C) Highly conserved glutamine residues Q625 (red) and Q629 (orange) project between the lobes and are mutated to arginine in human HELLS (identified in ICF syndrome proband E) and in Arabidopsis ddm1-9 (where it results in hypomethylation), respectively. The arginine residues are predicted to contact phosphates in the DNA minor groove and are also highlighted in (A). (D) A surface representation of the DDM1-nucleosome complex, highlighting the T80V mutation found in the male germline specific histone H3.3 MGH3 (red). T80 directly contacts DDM1 (Fig. 3C inset). (E) A surface representation of the DDM1-nucleosome complex, showing the surface exposed D382E mutation that results in a hypomethylation phenotype in ddm1-14. (F) A zoomed in view of the refined cryoEM density map, showing the N-terminal tail of histone H4 extending into the density observed for the DEXD ATPase domain of DDM1. Color coding of histone variants and DDM1 as in Figure 3.
