Figure 5. Human-specific features of the PDGFRA locus and glioma risk.
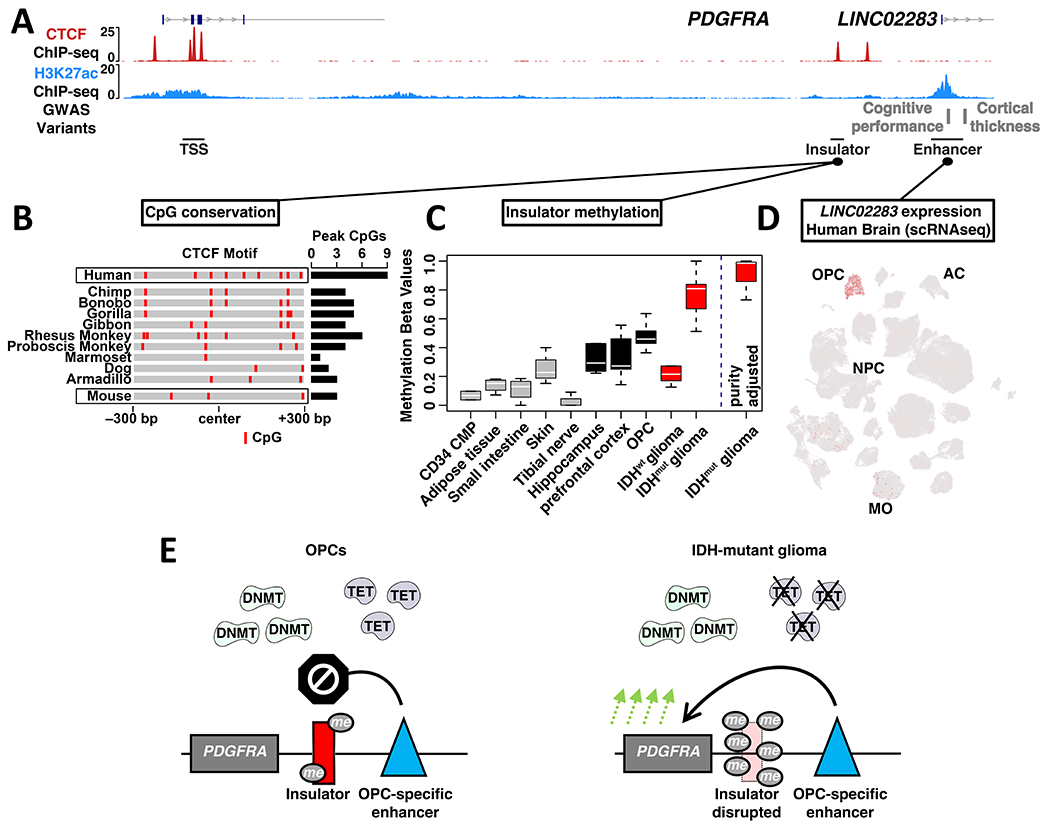
(A) ChIP-seq signals for CTCF (NPCs) and H3K27ac (OPCs) are shown for a ~110 kb region encompassing PDGFRA, insulator and OPC-specific enhancer. Genetic variants associated with cognitive performance and cortical thickness that coincide with the enhancer are indicated (GWAS), along with a corresponding non-coding RNA. (B) Heat depicts CpG dinucleotides over the PDGFRA insulator across representative mammalian species. Insulator intervals were defined based on synteny and conservation to the 600 bp CTCF binding peak called from human ChIP-seq data. The total number of CpGs in the 600 bp intervals is indicated for each species at right. (C) Box plot depicts PDGFRA insulator methylation in selected non-brain human cell and tissue types, human brain compartments, OPC-enriched fractions from human brain, and for IDHwt and IDHmut gliomas. Box plot at right depicts insulator methylation in IDHmut gliomas after correction for sample purity. (D) t-SNE plot generated from scRNA-seq of human brain cells in the middle temporal gyrus is annotated for expression of the non-coding RNA that emanates from the enhancer (LINC02283; MO: mature oligodendrocytes; AC: astrocytes; data from Allen brain map)50. (E) Proposed model contrasts insulator states in normal human OPCs and IDHmut glioma. In OPCs, methyltransferases (DNMT) and demethylases (TET) maintain low to intermediate methylation levels that leave the PDGFRA insulator largely intact. In IDHmut gliomas, inhibition of the demethylases results in hypermethylation and disruption of the insulator, driving aberrant PDGFRA expression and tumorigenesis.
