Fig. 1. Basic features of shark UrIg. A) UrIg is an ancient VJ domain.
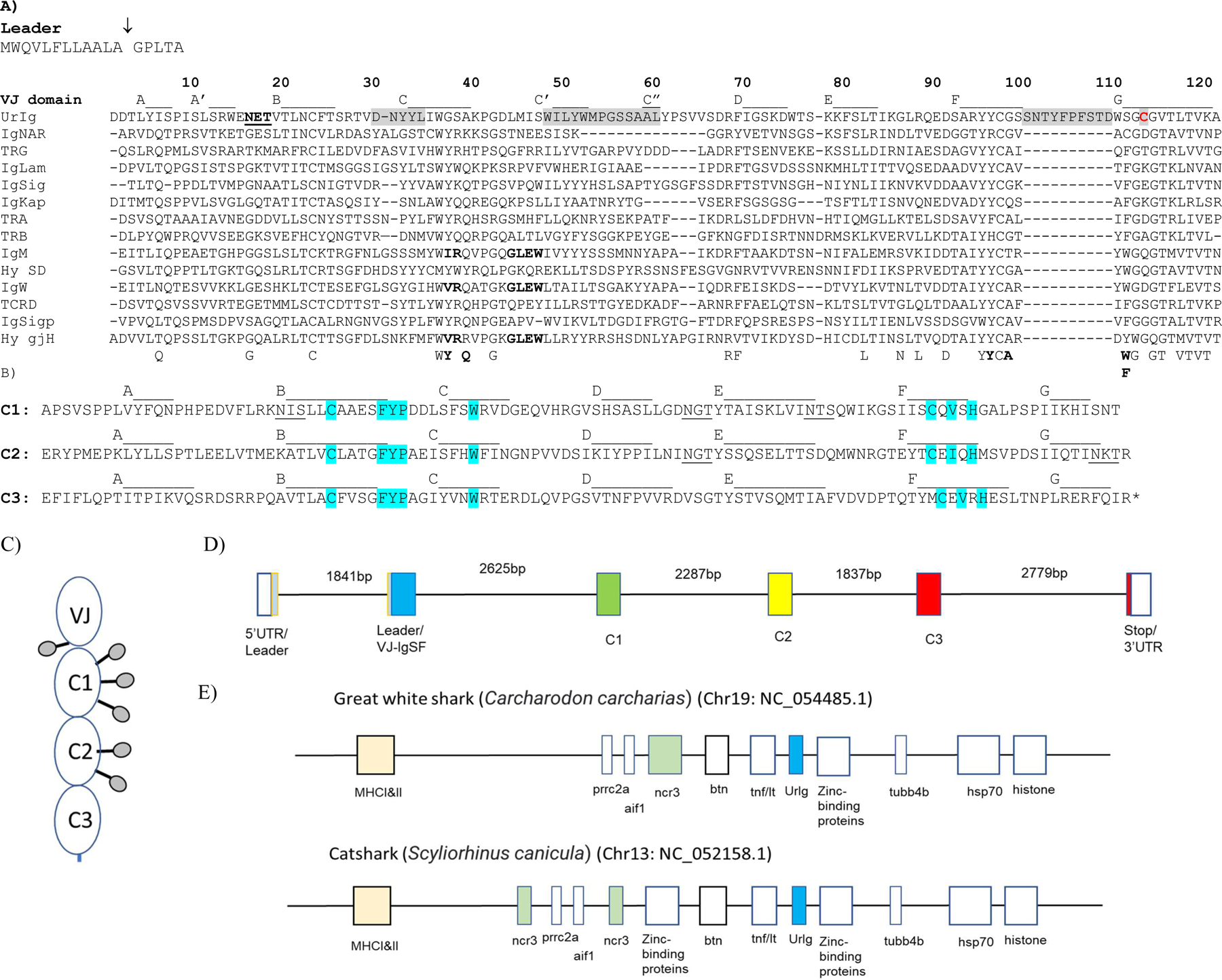
UrIg VJ domain aligned to nurse shark antigen receptor VJ domains: 4 types of TCR (α, β, γ, δ), IgM, IgW, IgNAR, and the 3 types of L chains (κ, λ, σ); the germline-joined Hydrolagus IgM V (Hyco-gjH) and single domain Hydrolagus IgM V (hyco-SD) are also aligned. Numbering and strand positions are based on the nurse shark UrIg mature protein sequence and AlphaFold prediction. Accession numbers for sequences are Nurse shark- UrIg (GIWU01140354.1); IgM (AAA50817); IgW (AAB08972): igNAR (AAB42621); IgLambda (sequence taken from; IgKappa (L16765); IgSigma (EF114759); TCRα (ADW95866); TCRβ (ADW95893): TCRδ (ADW95945); TCRγ (QNT23252). Spotted Ratfish (Hydrolagus colliei)- Hyco-SD (HHC1; seq taken from (63); Hyco-gjV (AAC12891). Conserved amino acids in VJ domains are shown below the figure. Residues that interact between V domains of bona fide Ig are bolded below the figure and in the IgM, IgW, and Hydrolagus V sequences. The potential glycosylation site in UrIg is underlined. CDR1, 2, and 3 are shaded. Only the CDR3 for UrIg is shown because the other CDR3s for Ig/TCR are somatically generated. The one free cysteine (Cys-113) in UrIg is shaded red. The arrow marking the leader segment indicates the intron of the “split leader.” Dashes indicate gaps in amino acid sequences. B) UrIg C domains bear classical C1 domain features found in Ig/TCR/MHC. The three C domains of UrIg are noted with the conserved residues in blue (esp. FYP and C-V-H). Strand positions are based on the AlphaFold prediction). Potential glycosylation sites are underlined. C. Schematic model of the UrIg monomer. Potential glycosylation sites shown as lollipops. D. Genomic (Intron/Exon) organization of nurse shark UrIg. Genomic DNA (position: 341176–354343 in JAHRHZ010000005) was aligned against a full-length transcript GIWU01140354 using splign. Each colored box is a coding region, and the untranslated regions (UTR) are in an open box. E. UrIg maps to the shark MHC in the class III region. UrIg, blue; the other VJ gene in the MHC, NCR3, in green; class I/II, light peach; other genes including class III genes are in open boxes. Mapping is not to scale, but simply shows relative locations. Wider boxes indicate clusters of multigenes.
