Fig. 2. Phylogenetic tree analyses of UrIg domains demonstrate UrIg’s ancient origin.
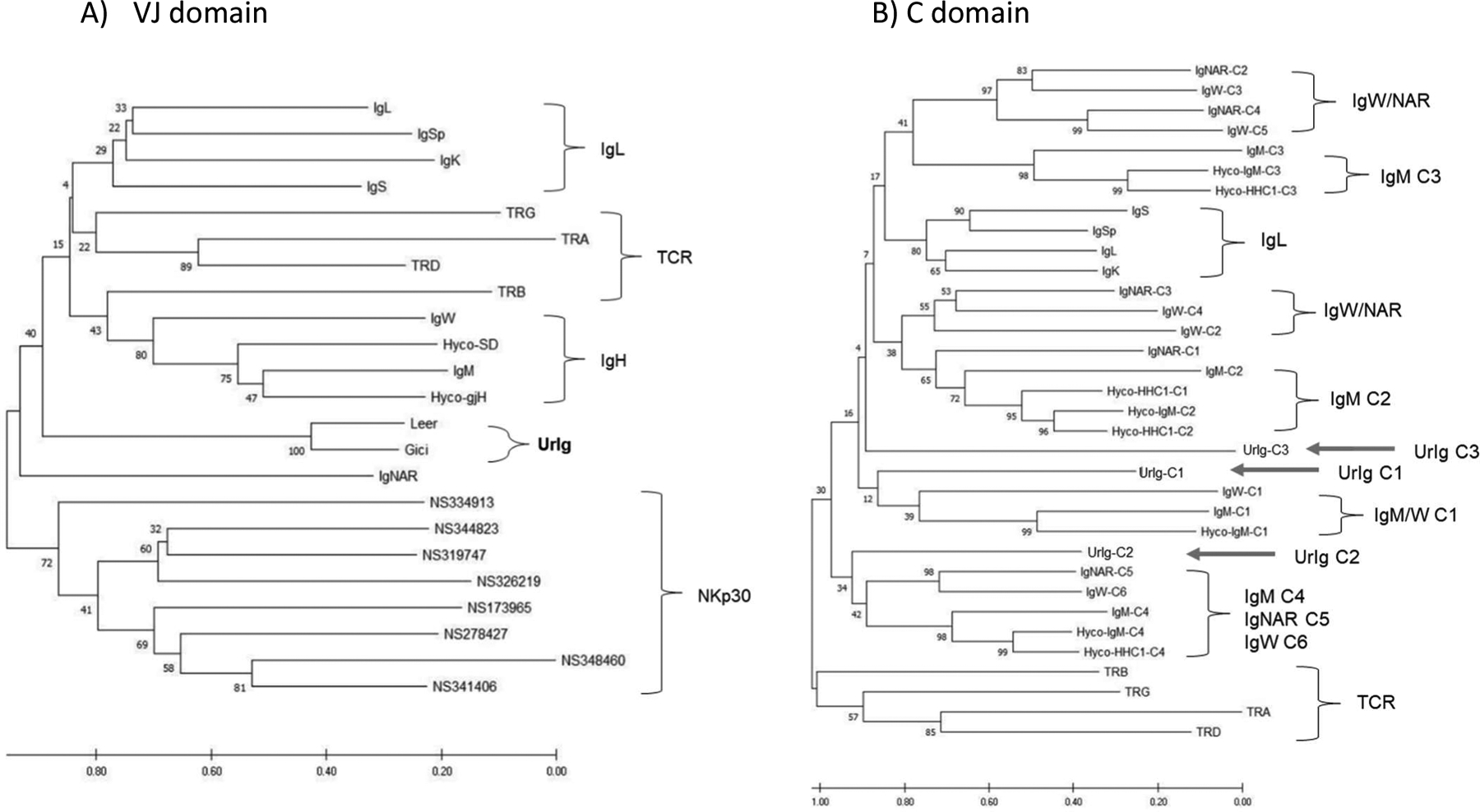
Neighbor-joining phylogenetic tree was performed with 500 bootstrap runs. The original tree is shown. Scale bars indicate the number of amino acid changes per site. A. Tree of VJ domains from nurse shark Ig/TCR and NKp30. UrIg VJ from two highly divergent Elasmobranchs nurse shark Ginglymostoma cirratum (Gici) and little skate Leucoraja erinacea (Leer) included. Ratfish Hydrolagus colliei IgM (Hyco-SD) and germline-joined IgM (Hyco-gH) included. CDR3 were removed from the sequences to construct the tree. B. Relationships of UrIg C domains to nurse shark Ig/TCR (and Hydrolagus IgM) C domains. Positions of the three UrIg C domains shown with arrows. Since clustering was more informative, we did not root these trees. Additional accession numbers for these analyses are: V- nurse shark IgS’ (Sp) (AAV34678); Nurse shark NCR3 genes (MT914154–914161); C- Hyco-IgM (AAC12891); TCRα (ADW95866; JAHRHZ010000115.1); TCRβ (ADW95903): TCRδ (ADW95942); TCRγ (ADW95913).
