Figure 1. The architecture of MutL homologs promotes DNA loading by MutS homologs.
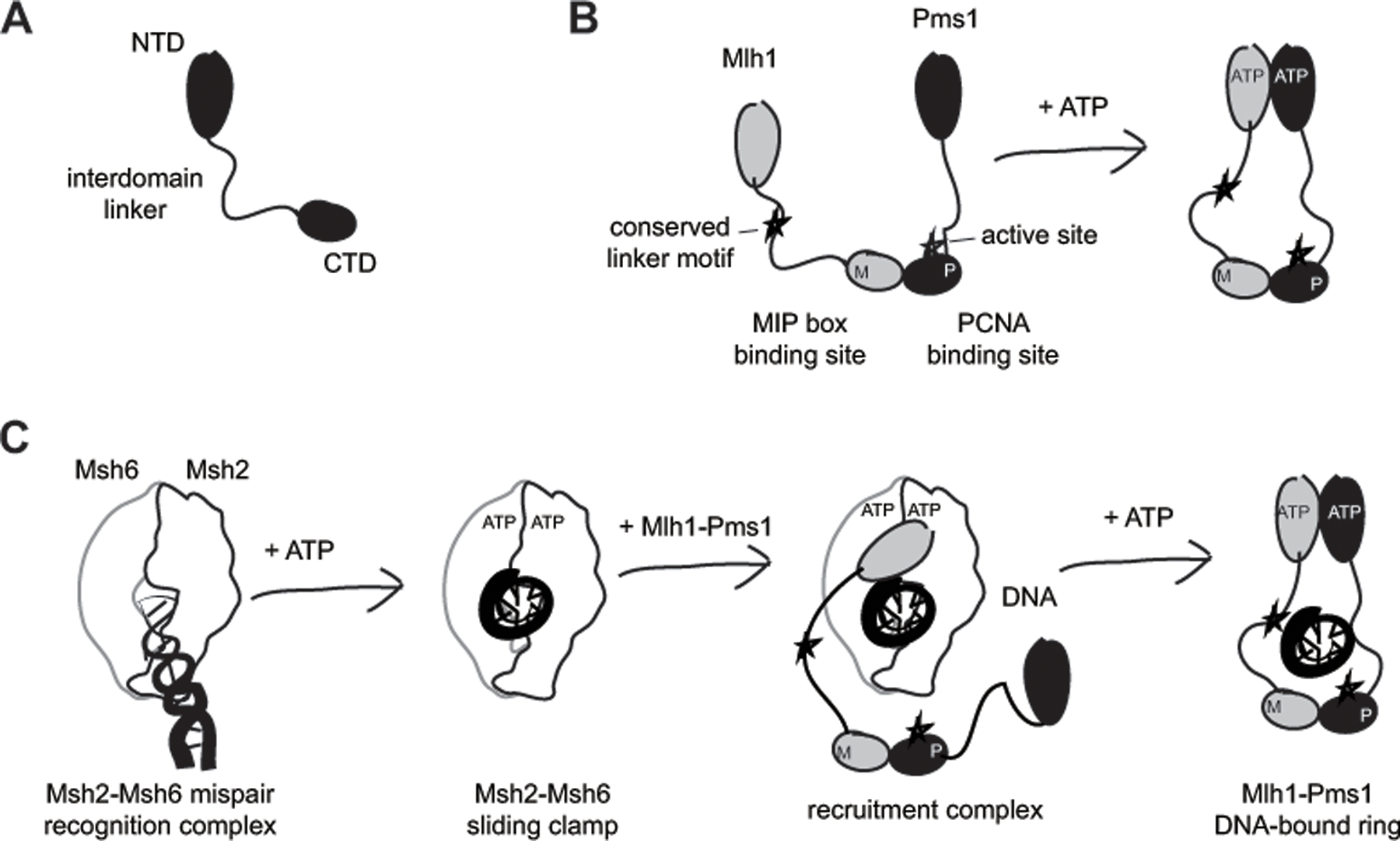
A. MutL homologs can be divided into an N-terminal domain (NTD) belonging to the GHKL ATPase family, a C-terminal domain (CTD) and an unstructured interdomain linker. B. The CTDs are constitutively dimerized, whereas the NTD dimerization is driven by ATP binding. In the heterodimeric S. cerevisiae Mlh1-Pms1, the Mlh1 subunit lacks the CTD endonuclease active site and PCNA binding site, but has a site for binding MIP (Mlh1 Interacting Peptide) box proteins and a conserved linker motif required for efficient DNA nicking. C. DNA loading involves sequential mispair and ATP binding by Msh2-Msh6 to form a sliding clamp that is capable of recruiting Mlh1-Pms1 to DNA. Mlh1-Pms1 ring formation is mediated by Mlh1 NTD binding by Msh2-Msh6[17] and ATP-mediated dimerization of the Mlh1 and Pms1 NTDs,[5] and is facilitated by the flexible interdomain linker.
