Figure 2. The Mlh1 conserved linker motif is conserved with two sequence patterns in eukaryotes.
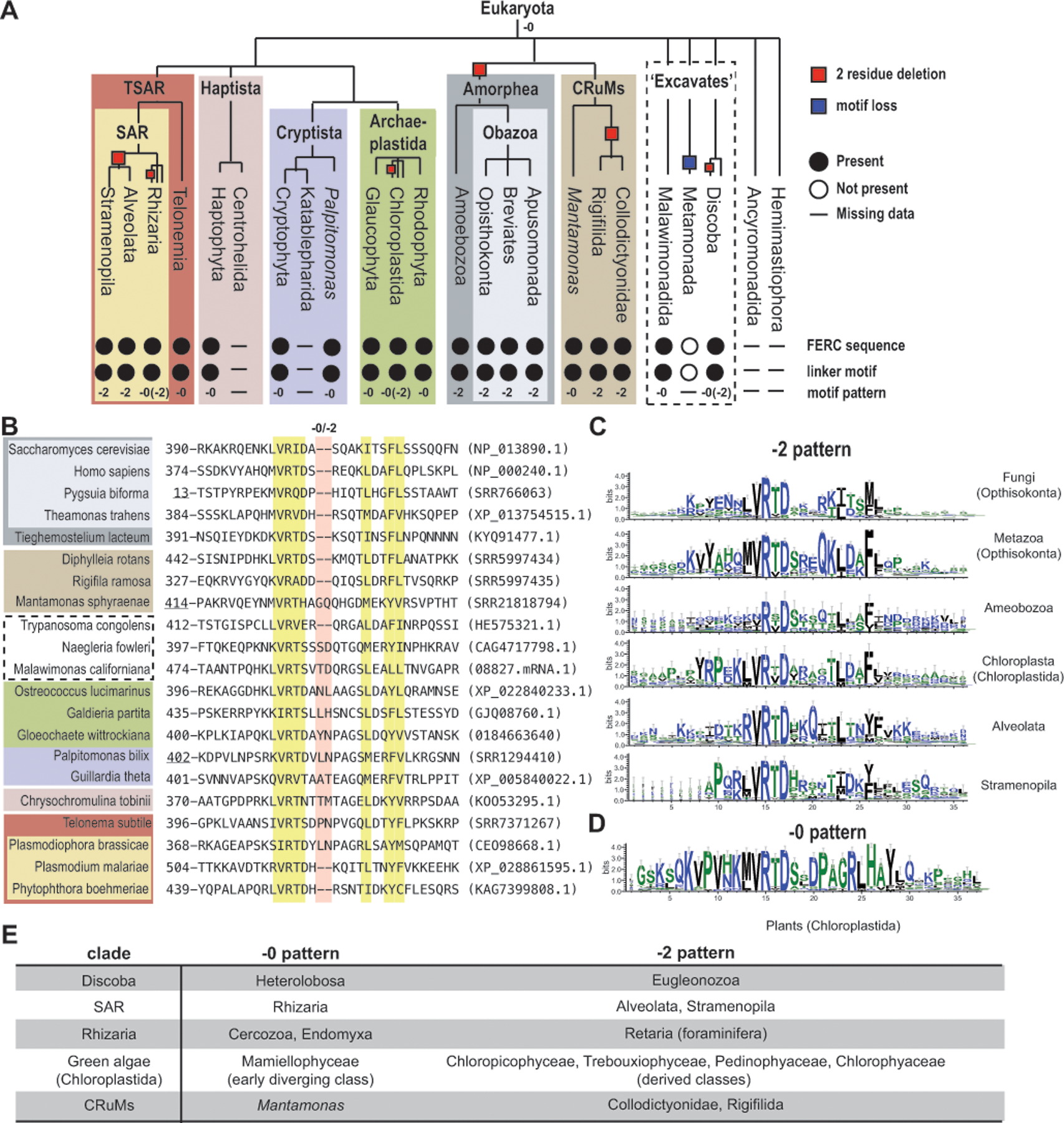
A. The presence (filled circles), absence (empty circles), or no data (horizontal line) of the conserved Mlh1 linker motif and C-terminal FERC sequence in different eukaryotic clades is indicated; the −0 and −2 labels indicate which motif pattern is present. The cladogram is based on previous analyses.[10] B. The conserved linker motif from the alignment of selected Mlh1 sequences; yellow highlighted residues are conserved and red highlighted residues are at −0/−2 sequence difference. Accessions are in parentheses with SRR numbers corresponding to RNA sequence datasets; Mlh1 sequences from other sources include M. californiana (https://megasun.bch.umontreal.ca/papers/T2SS-2020/ sequenceMalawimonas_californiana_08827.mRNA.1), G. wittrockiana (MMETSP0308 sequence CAMPEP_0184663640), C. tobinii (KOO53295.1 where two in-frame introns were merged with the coding sequence). Underlined positions are from partial sequences. C-D. Motif sequence logos were generated with Weblogo3[14]. E. Clades with both −0 and −2 patterns have patterns that segregate by taxonomic group.
