Figure 3. Linker peptide conformations do not explain sequence constraints.
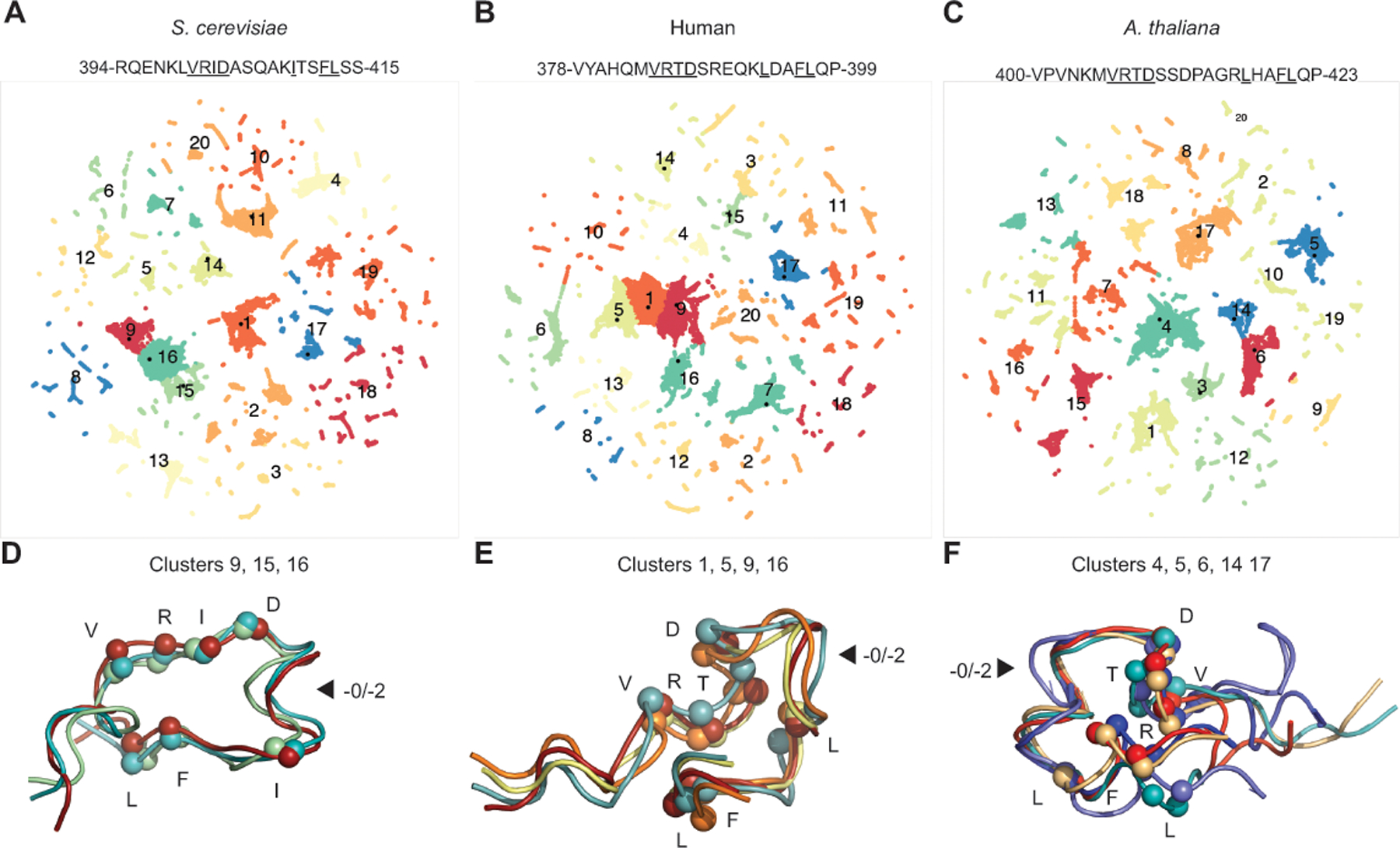
A-C. Mlh1 motif-containing peptides were analyzed by molecular dynamics and related conformations (clusters) were identified by two-dimensional UMAP projections. D-F. The most common conformations are displayed as ribbons with Cα atoms of the conserved residues (underlined in sequence) displayed as spheres. Arrows indicate where the length difference between the −0 and −2 patterns lies. D. The major S. cerevisiae peptide conformation is stabilized by interactions between R401, I402, and F412 and capping of a helix with the D403 side chain. E. The major human peptide conformation is stabilized by interactions between 384-VRTD-387, L393, and 396-FL-397. F. The major A. thaliana peptide conformation contains a helix promoted by the non-conserved P413 and is stabilized by interactions between R407, L417, and F420.
