Figure 5. The conserved linker likely interacts with the DNA substrate.
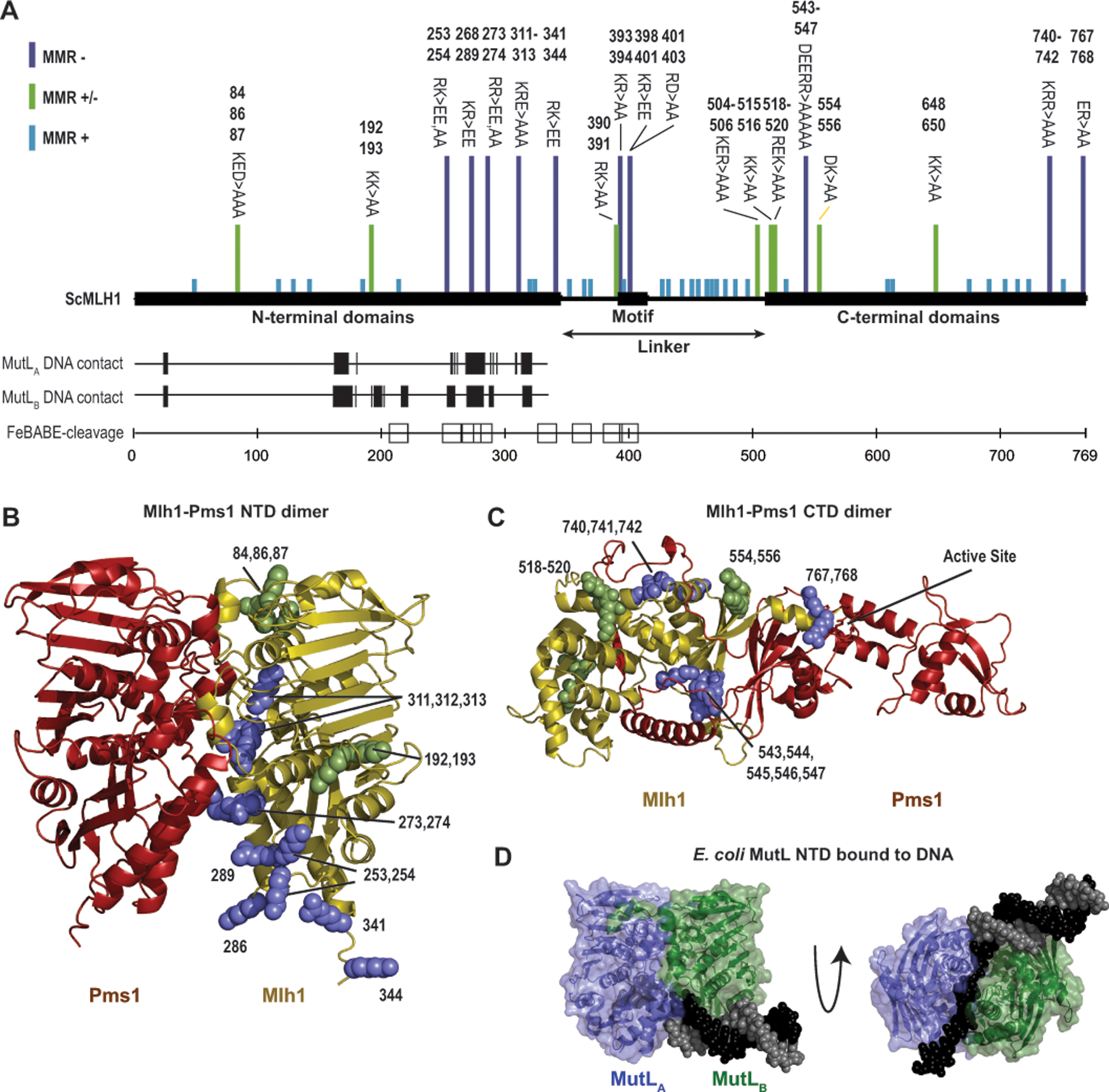
A. Summary of the data indicating regions of S. cerevisiae Mlh1 that interact with DNA provide a consistent view of DNA-interacting residues. Bars above indicate mutations causing full (purple), partial (green) or no (blue) MMR defects.[4, 13, 37, 73] Vertical hashes indicate residues equivalent to DNA contacting residues in each subunit (MutLA and MutLB) of the E. coli MutL-DNA structure.[7] Sites of S. cerevisiae Mlh1 adjacent to DNA in a Mlh1-Mlh3 bound to a Holliday junction[13] are indicated by squares. B. Sites of mutations in Mlh1 that cause complete (purple) or partial (green) MMR defects mapped to a Mlh1-Pms1 NTD dimer model generated with Alphafold2.[40] C. Similar analysis as panel B with a Mlh1-Pms1 CTD dimer model. D. Structure of E. coli MutL (green and blue cartoon) with a DNA containing a single-stranded overhang (black and gray spheres) shows extensive ssDNA-MutL NTD contacts (PDB id 7p8v).[7]
