Figure 6. The active site of eukaryotic Mlh1-Pms1 and Mlh1-Mlh3 is blocked by the Mlh1 C-terminus.
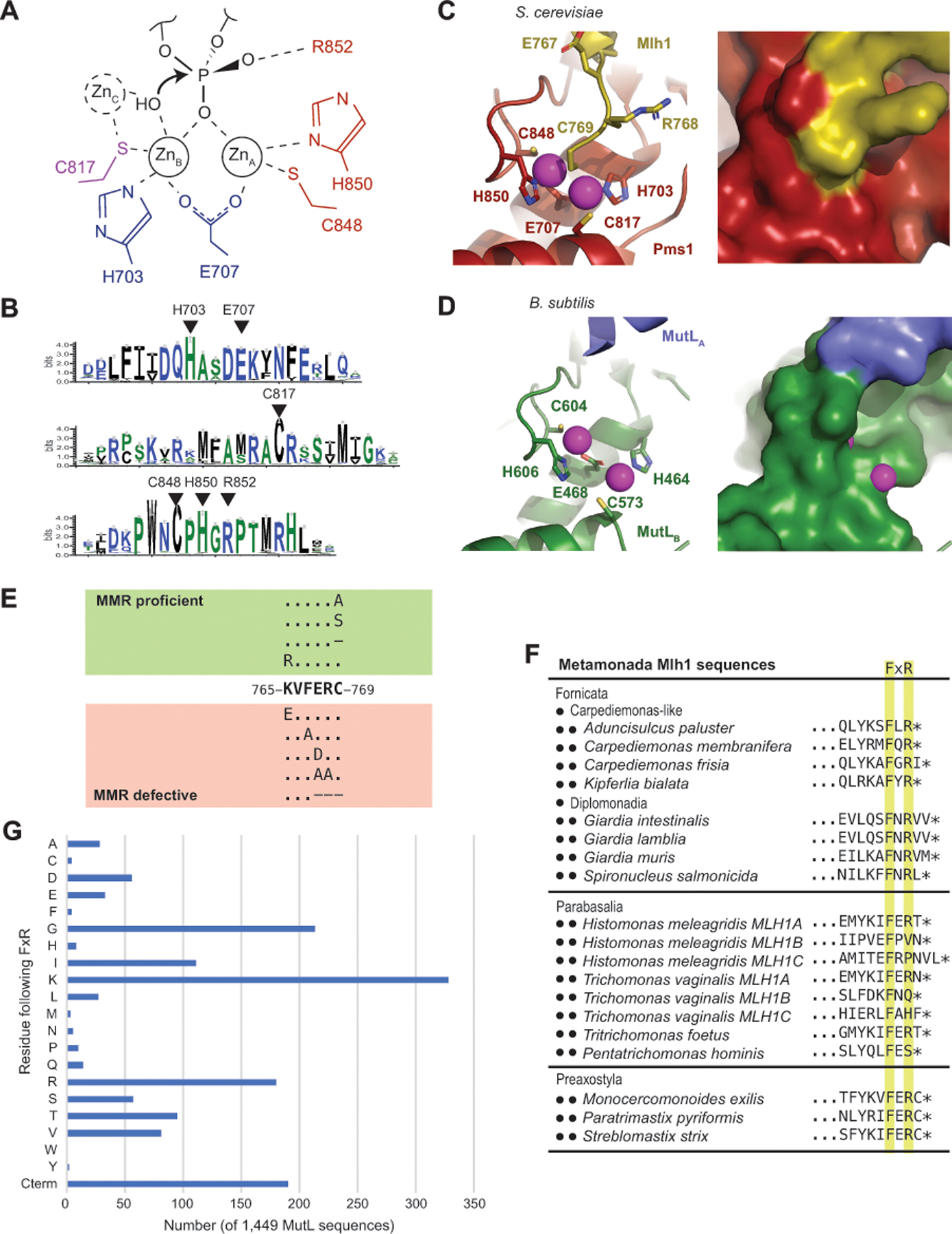
A. The proposed endonuclease reaction of MutL-family proteins involves coordination of a backbone phosphate by ZnA and ZnB, stabilization of the phosphate by Pms1 R852 and potential activation of a hydroxyl nucleophile by a transiently bound ZnC; residues identified with S. cerevisiae Pms1 numbering. B. Sequence logo of the conserved active site motifs in eukaryotic Pms1 sequences with residues identified in panel A indicated with triangles. C. Ribbon diagrams (left and middle) of the S. cerevisiae Mlh1 (yellow) – Pms1 (red) active site (PDB id 4e4w; [34]) showing the metal coordination and binding of the Mlh1 C769 residue at the bridging position between ZnA and ZnB (magenta). Surface view of the active site shows that not only is the fourth coordination sphere blocked by C769, but the entire active site is sequestered from solvent. D. Ribbon diagram (left) and surface (right) of the B. subtilis active site (PDB id 3kdk; [63]) showing accessibility of the metals (magenta) to solvent. E. Summary of the mutagenesis of the conserved S. cerevisiae Mlh1 C-terminus; dashes indicate deletions of the specified residue. F. Two of the three phyla within Metamonada lack the C-terminal Mlh1 FERC sequence but tend to retain a FxR sequence. G. Histogram of the residues that immediately follow the FxR sequence from a sample of 1,449 bacterial and archaeal MutL sequences shows a bias towards small and positively charged residues and a relative lack of cystine residues as in the eukaryotic Mlh1 FERC motif. “Cterm” indicates that the arginine in the FxR sequence is at the C-terminus of the protein.
