Figure 2. Chromosomal duplications containing murZ or murA are present in ΔgpsB or ΔstkP suppressor strains of S. pneumoniae D39.
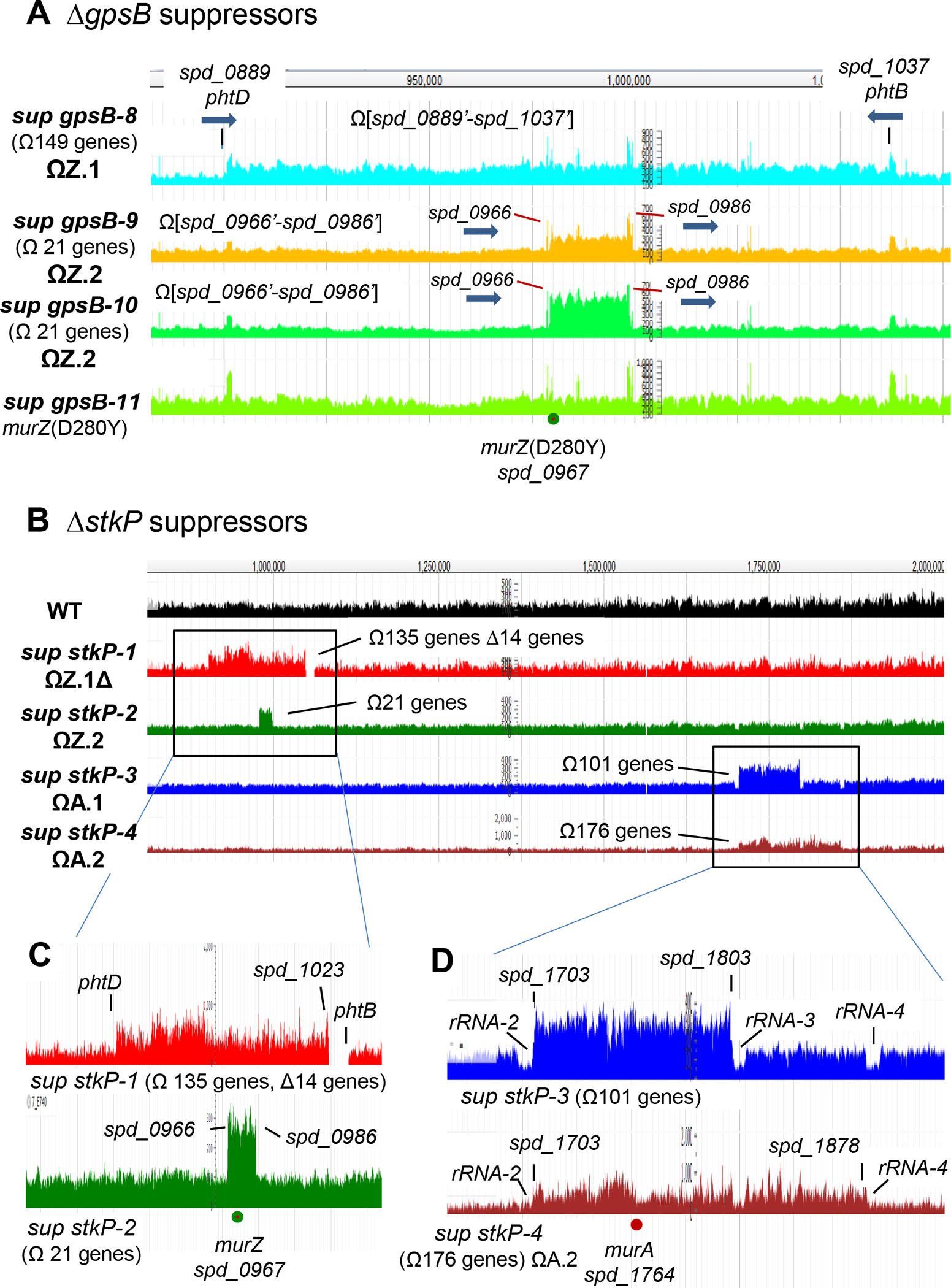
(A) Snapshot of genome browser output of ΔgpsB sup strains from genome coordinates 870 to 1100 kb. Three new ΔgpsB suppressor strains contain chromosomal duplication or quadruplication of multiple genes, all of which include murZ. Sup gpsB-8 contains a ≈163 kb duplication of chromosomal region from spd_0889’ to spd_1037’, while sup gpsB-9 and sup gpsB-10 contain a duplication or quadruplication, respectively, of the chromosomal region from spd_0966’ to spd_0986’. sup gpsB-10 has a murZ(D280Y) mutation and no chromosomal duplication. Black lines point to the flanking regions of the duplication found in sup gpsB-8, which are 1324-bp inverted repeats present in phtD (spd_0889) and phtB (spd_1037), encoding 2 of the 3 pneumococcal histidine triad proteins. The red lines point to the flanking regions (spd_0966 and spd_0986) of duplication or quadruplication found in sup gpsB-9, and -10, respectively. spd_0966 and spd_0986 are pseudogenes containing IS1167 degenerate transposase sequences. Thick blue arrows show the gene orientations of phtB, phtD, spd_0966, and spd_0986. (B) Snapshot of genome browser output of ΔstkP sup strains from genome coordinates 750 to 2,000 kb. (C) Sup stkP-1 contains a duplication/deletion between phtD and phtB, and sup stkP-2 contains a duplication between spd_0966 and spd_0986. (D) Large duplications found in sup stkP-3 and -4 are flanked by tRNA + rRNA clusters rRNA/rRNA3 and rRNA/rRNA4 respectively. Sup stkP-3 showed a decrease in sequence reads of the four rRNA-1–4 operons (rRNA-1, rRNA-2, rRNA-3, and rRNA-4) compared to the surrounding region. It is possible that either rRNA-2 or rRNA-3, or both rRNA-2 and rRNA-3, are deleted in this strain, but because of the sequence identity of the rRNA operons, deletion of one or two operons manifest as a decrease of reads for all four operons.
